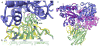Allosteric control of ligand-binding affinity using engineered conformation-specific effector proteins - PubMed (original) (raw)
Allosteric control of ligand-binding affinity using engineered conformation-specific effector proteins
Shahir S Rizk et al. Nat Struct Mol Biol. 2011 Apr.
Abstract
We describe a phage display methodology for engineering synthetic antigen binders (sABs) that recognize either the apo or the ligand-bound conformation of maltose-binding protein (MBP). sABs that preferentially recognize the maltose-bound form of MBP act as positive allosteric effectors by substantially increasing the affinity for maltose. A crystal structure of a sAB bound to the closed form of MBP reveals the basis for this allosteric effect. We show that sABs that recognize the bound form of MBP can rescue the function of a binding-deficient mutant by restoring its natural affinity for maltose. Furthermore, the sABs can enhance maltose binding in vivo, as they provide a growth advantage to bacteria under low-maltose conditions. The results demonstrate that structure-specific sABs can be engineered to dynamically control ligand-binding affinities by modulating the transition between different conformations.
Figures
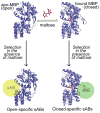
Figure 1. Phage display selection strategy
Maltose binding protein (MBP) undergoes a conformational change through a hinge-bending motion upon binding to maltose (red). Carrying out the phage display selection in the absence of maltose generates sABs (yellow sphere) that bind preferentially to the open form of MBP, whereas selection in the presence of maltose results in closed-specific sABs (green sphere). Note: placement of spheres indicate postulated binding modes of sABs to the different forms of MBP.
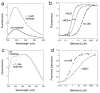
Figure 2. Influence of sABs on maltose binding
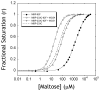
(A) The change in fluorescence of the MBP-233C Alexa 488 conjugate (solid line) upon addition of 1 mM maltose (dashed line). (B) Fluorescence maltose binding curves of MBP-233C Alexa 488 in the absence (●) or presence of 200 nM sAB MCS1 (◇) or 200 nM sAB MCS4 (○). (C) Intrinsic tryptophan fluorescence of MBP in the absence (solid line) or presence of 1 mM maltose (dashed line). (D) Fluorescence maltose binding curves of MBP the absence (●) or presence of 200 nM sAB MOS1 (○).
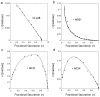
Figure 3. Scatchard analysis of maltose binding
Data points from figure 2 were re-plotted as r vs. r/[Maltose], where r is the fractional saturation of MBP with maltose. (A) Binding of maltose to MBP in the absence of sABs showing no co-operativity. (B) Maltose binding in the presence of 200 nM sAB MOS1 showing negative co-operativity. (C) Maltose binding in the presence of 200 nM sAB MCS1 or (D) 200 nM sAB MCS4 showing positive co-operativity.
Figure 4. Crystal structure of MBP-MCS2 complex
MBP (blue) is in the closed form, bound to 1 molecule of maltose (red). The sAB (Heavy chain: green, Light chain: yellow) interacts with MBP at the opposite side of the binding pocket forming a wedge that favors the closed form.
Figure 5. The “wedge” formed by the CDR loops of MCS2
(A) A group of bulky side-chain residues within CDRH-3 of the sAB (green sticks) form the wedge structure, which interacts with a region within MBP (blue) that is only exposed in the closed, maltose-bound conformation. (B) Sequence of the CDR loops of MCS2. Bold letters indicate residues that interact with the MBP molecule. (C) An overlay of the MCS2-MBP complex with the open form of MBP (purple, PDB code: 1OMP 12) indicates that the apo form of MBP clashes with the sAB CDR loops.
Figure 6. Rescuing binding function of an MBP mutant
The effect of the binding pocket mutation W62F on affinity of MBP for maltose was assessed using the emission change of Alexa 488 attached to a cysteine at position 233. In the absence of sAB (●), the mutant exhibits low binding affinity. The affinity is restored by addition of either 200 nM sAB MCS4 (○) or 200 nM sAB MCS1 (◇). The binding curve of MBP-233C with no binding pocket mutations is shown (dashed line) as a reference.
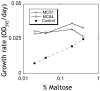
Figure 7. Alloesteric activity of sABs in vivo
E. coli cells expressing sABs in the periplasm were grown in minimal media containing maltose as the sole carbon source. Cells expressing sAB MCS1 (○) or sAB MCS4 (◇) show no change in the growth rate at low maltose concentrations. Control cells expressing sAB-27 (■), which is an actin binding sAB (from reference14), show a lower growth rate at low maltose concentrations.
Similar articles
- Engineered synthetic antibodies as probes to quantify the energetic contributions of ligand binding to conformational changes in proteins.
Mukherjee S, Griffin DH, Horn JR, Rizk SS, Nocula-Lugowska M, Malmqvist M, Kim SS, Kossiakoff AA. Mukherjee S, et al. J Biol Chem. 2018 Feb 23;293(8):2815-2828. doi: 10.1074/jbc.RA117.000656. Epub 2018 Jan 10. J Biol Chem. 2018. PMID: 29321208 Free PMC article. - Modulation of effector affinity by hinge region mutations also modulates switching activity in an engineered allosteric TEM1 beta-lactamase switch.
Kim JR, Ostermeier M. Kim JR, et al. Arch Biochem Biophys. 2006 Feb 1;446(1):44-51. doi: 10.1016/j.abb.2005.11.014. Epub 2005 Dec 9. Arch Biochem Biophys. 2006. PMID: 16384549 - Manipulation of ligand binding affinity by exploitation of conformational coupling.
Marvin JS, Hellinga HW. Marvin JS, et al. Nat Struct Biol. 2001 Sep;8(9):795-8. doi: 10.1038/nsb0901-795. Nat Struct Biol. 2001. PMID: 11524684 - The dynamics of the MBP-MalFGK(2) interaction: a prototype for binding protein dependent ABC-transporter systems.
Shilton BH. Shilton BH. Biochim Biophys Acta. 2008 Sep;1778(9):1772-80. doi: 10.1016/j.bbamem.2007.09.005. Epub 2007 Sep 19. Biochim Biophys Acta. 2008. PMID: 17950243 Review. - Structures of a hemoglobin-based blood substitute: insights into the function of allosteric proteins.
Kroeger KS, Kundrot CE. Kroeger KS, et al. Structure. 1997 Feb 15;5(2):227-37. doi: 10.1016/s0969-2126(97)00181-0. Structure. 1997. PMID: 9032082 Review.
Cited by
- Conformational stabilization of ubiquitin yields potent and selective inhibitors of USP7.
Zhang Y, Zhou L, Rouge L, Phillips AH, Lam C, Liu P, Sandoval W, Helgason E, Murray JM, Wertz IE, Corn JE. Zhang Y, et al. Nat Chem Biol. 2013 Jan;9(1):51-8. doi: 10.1038/nchembio.1134. Epub 2012 Nov 25. Nat Chem Biol. 2013. PMID: 23178935 - Protein targeting. Structure of the Get3 targeting factor in complex with its membrane protein cargo.
Mateja A, Paduch M, Chang HY, Szydlowska A, Kossiakoff AA, Hegde RS, Keenan RJ. Mateja A, et al. Science. 2015 Mar 6;347(6226):1152-5. doi: 10.1126/science.1261671. Science. 2015. PMID: 25745174 Free PMC article. - An allosteric switch for pro-HGF/Met signaling using zymogen activator peptides.
Landgraf KE, Steffek M, Quan C, Tom J, Yu C, Santell L, Maun HR, Eigenbrot C, Lazarus RA. Landgraf KE, et al. Nat Chem Biol. 2014 Jul;10(7):567-73. doi: 10.1038/nchembio.1533. Epub 2014 May 25. Nat Chem Biol. 2014. PMID: 24859116 - A conformation-selective monoclonal antibody against a small molecule-stabilised signalling-deficient form of TNF.
Lightwood DJ, Munro RJ, Porter J, McMillan D, Carrington B, Turner A, Scott-Tucker A, Hickford ES, Schmidt A, Fox D 3rd, Maloney A, Ceska T, Bourne T, O'Connell J, Lawson ADG. Lightwood DJ, et al. Nat Commun. 2021 Jan 25;12(1):583. doi: 10.1038/s41467-020-20825-6. Nat Commun. 2021. PMID: 33495445 Free PMC article. - Antibody-enabled small-molecule drug discovery.
Lawson AD. Lawson AD. Nat Rev Drug Discov. 2012 Jun 29;11(7):519-25. doi: 10.1038/nrd3756. Nat Rev Drug Discov. 2012. PMID: 22743979 Review.
References
- Lowman HB, Wells JA. Affinity maturation of human growth hormone by monovalent phage display. J Mol Biol. 1993;234:564–78. - PubMed
- Monod J, Wyman J, Changeux JP. On the Nature of Allosteric Transitions: A Plausible Model. J Mol Biol. 1965;12:88–118. - PubMed
- Koshland DE, Jr, Nemethy G, Filmer D. Comparison of experimental binding data and theoretical models in proteins containing subunits. Biochemistry. 1966;5:365–85. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- GM 072688/GM/NIGMS NIH HHS/United States
- F32DK080619-02/DK/NIDDK NIH HHS/United States
- U01 GM094588/GM/NIGMS NIH HHS/United States
- R01 GM072688/GM/NIGMS NIH HHS/United States
- F32 DK080619/DK/NIDDK NIH HHS/United States
- F32 DK080619-03/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous