A bioinformatic assay for pluripotency in human cells - PubMed (original) (raw)
doi: 10.1038/nmeth.1580. Epub 2011 Mar 6.
Bernhard M Schuldt, Roy Williams, Dylan Mason, Gulsah Altun, Eirini P Papapetrou, Sandra Danner, Johanna E Goldmann, Arne Herbst, Nils O Schmidt, Josef B Aldenhoff, Louise C Laurent, Jeanne F Loring
Affiliations
- PMID: 21378979
- PMCID: PMC3265323
- DOI: 10.1038/nmeth.1580
A bioinformatic assay for pluripotency in human cells
Franz-Josef Müller et al. Nat Methods. 2011 Apr.
Abstract
Pluripotent stem cells (PSCs) are defined by their potential to generate all cell types of an organism. The standard assay for pluripotency of mouse PSCs is cell transmission through the germline, but for human PSCs researchers depend on indirect methods such as differentiation into teratomas in immunodeficient mice. Here we report PluriTest, a robust open-access bioinformatic assay of pluripotency in human cells based on their gene expression profiles.
Figures
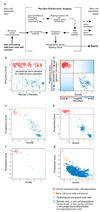
Figure 1. A multidimensional data model for assessing pluripotent stem cells
A Schematic for PluriTest. (b–c) We constructed and optimized a multi-class classifier (Pluripotency Score, b, c) and a one-class classifier (Novelty Score, c) to distinguish pluripotent stem cells (PSC) from other cell types and tissues. In (b), we show pluripotent (red) and somatic samples (blue) in the training dataset as assessed with the Pluripotency Score and in (c) with both PluriTest scores. In (d–g), we plot Pluripotency Scores against Novelty Scores for test data set samples. The classifiers were tested against datasets generated on four different microarray platforms: Illumina WG6v1 (c, 177 samples), HT12v3 (d, 498 samples), HT12v4 (e, 38 samples) and Affymetrix U133A (f, 5372 samples)10). Samples for these datasets were independently generated (c and d) or curated from published studies (c, d, f). In d, the lines in the plot indicate empirically determined thresholds for defining normal pluripotent lines (see also Supplementary Fig. 2).
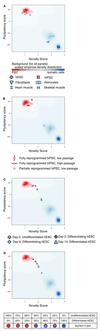
Figure 2. Application of PluriTest
The graphs show the actual output of PluriTest. Pluripotency score is plotted against novelty score for the indicated samples. The background encodes an empirical density map indicating pluripotency (red) and novelty (blue). (a–c) PluriTest results for known pluripotent cells and somatic cells and tissues (a), for fully and partially reprogrammed iPSC lines (b), and for an hESC line (WA09) being differentiated into neural precursors, at the indicated time points. In (d) PluriTest was run on mixed samples of hESC and hESC-derived neural precursor RNA (day 0 and day 14 from c) at the indicated ratios. hESC, human embryonic stem cell, hiPSC, human induced pluripotent stem cell.
Comment in
- Testing pluripotency.
de Souza N. de Souza N. Nat Methods. 2011 Apr;8(4):287. doi: 10.1038/nmeth0411-287. Nat Methods. 2011. PMID: 21574272 No abstract available.
Similar articles
- A guide to stem cell identification: progress and challenges in system-wide predictive testing with complex biomarkers.
Williams R, Schuldt B, Müller FJ. Williams R, et al. Bioessays. 2011 Nov;33(11):880-90. doi: 10.1002/bies.201100073. Epub 2011 Sep 8. Bioessays. 2011. PMID: 21901750 Review. - Assessment of established techniques to determine developmental and malignant potential of human pluripotent stem cells.
International Stem Cell Initiative. International Stem Cell Initiative. Nat Commun. 2018 May 15;9(1):1925. doi: 10.1038/s41467-018-04011-3. Nat Commun. 2018. PMID: 29765017 Free PMC article. - Gene regulation networks related to neural differentiation of hESC.
Zhong JF, Song Y, Du J, Gamache C, Burke KA, Lund BT, Weiner LP. Zhong JF, et al. Gene Expr. 2007;14(1):23-34. doi: 10.3727/000000007783991781. Gene Expr. 2007. PMID: 17933216 Free PMC article. - Regulatory networks define phenotypic classes of human stem cell lines.
Müller FJ, Laurent LC, Kostka D, Ulitsky I, Williams R, Lu C, Park IH, Rao MS, Shamir R, Schwartz PH, Schmidt NO, Loring JF. Müller FJ, et al. Nature. 2008 Sep 18;455(7211):401-5. doi: 10.1038/nature07213. Epub 2008 Aug 24. Nature. 2008. PMID: 18724358 Free PMC article. - The cell cycle and pluripotency.
Hindley C, Philpott A. Hindley C, et al. Biochem J. 2013 Apr 15;451(2):135-43. doi: 10.1042/BJ20121627. Biochem J. 2013. PMID: 23535166 Free PMC article. Review.
Cited by
- A quantitative system for discriminating induced pluripotent stem cells, embryonic stem cells and somatic cells.
Wang A, Du Y, He Q, Zhou C. Wang A, et al. PLoS One. 2013;8(2):e56095. doi: 10.1371/journal.pone.0056095. Epub 2013 Feb 13. PLoS One. 2013. PMID: 23418520 Free PMC article. - Small molecules enable highly efficient neuronal conversion of human fibroblasts.
Ladewig J, Mertens J, Kesavan J, Doerr J, Poppe D, Glaue F, Herms S, Wernet P, Kögler G, Müller FJ, Koch P, Brüstle O. Ladewig J, et al. Nat Methods. 2012 Jun;9(6):575-8. doi: 10.1038/nmeth.1972. Epub 2012 Apr 8. Nat Methods. 2012. PMID: 22484851 - Human Cell Atlas and cell-type authentication for regenerative medicine.
Panina Y, Karagiannis P, Kurtz A, Stacey GN, Fujibuchi W. Panina Y, et al. Exp Mol Med. 2020 Sep;52(9):1443-1451. doi: 10.1038/s12276-020-0421-1. Epub 2020 Sep 15. Exp Mol Med. 2020. PMID: 32929224 Free PMC article. Review. - TeratoScore: Assessing the Differentiation Potential of Human Pluripotent Stem Cells by Quantitative Expression Analysis of Teratomas.
Avior Y, Biancotti JC, Benvenisty N. Avior Y, et al. Stem Cell Reports. 2015 Jun 9;4(6):967-74. doi: 10.1016/j.stemcr.2015.05.006. Stem Cell Reports. 2015. PMID: 26070610 Free PMC article. - Opportunities and challenges of pluripotent stem cell neurodegenerative disease models.
Sandoe J, Eggan K. Sandoe J, et al. Nat Neurosci. 2013 Jul;16(7):780-9. doi: 10.1038/nn.3425. Epub 2013 Jun 25. Nat Neurosci. 2013. PMID: 23799470 Review.
References
- Daley GQ, Lensch MW, Jaenisch R, et al. Cell Stem Cell. 2009;4(3):200. - PubMed
- Takahashi K, Yamanaka S. Cell. 2006;126(4):663. - PubMed
- Müller FJ, Goldmann J, Loser P, et al. Cell Stem Cell. 2010;6(5):412. - PubMed
- Russell WMS, Burch RL. The Principles of Humane Experimental Technique. London: Methuen; 1959.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials