A rare variant in MYH6 is associated with high risk of sick sinus syndrome - PubMed (original) (raw)
. 2011 Mar 6;43(4):316-20.
doi: 10.1038/ng.781.
Daniel F Gudbjartsson, Patrick Sulem, Gisli Masson, Hafdis Th Helgadottir, Carlo Zanon, Olafur Th Magnusson, Agnar Helgason, Jona Saemundsdottir, Arnaldur Gylfason, Hrafnhildur Stefansdottir, Solveig Gretarsdottir, Stefan E Matthiasson, Gu Mundur Thorgeirsson, Aslaug Jonasdottir, Asgeir Sigurdsson, Hreinn Stefansson, Thomas Werge, Thorunn Rafnar, Lambertus A Kiemeney, Babar Parvez, Raafia Muhammad, Dan M Roden, Dawood Darbar, Gudmar Thorleifsson, G Bragi Walters, Augustine Kong, Unnur Thorsteinsdottir, David O Arnar, Kari Stefansson
Affiliations
- PMID: 21378987
- PMCID: PMC3066272
- DOI: 10.1038/ng.781
A rare variant in MYH6 is associated with high risk of sick sinus syndrome
Hilma Holm et al. Nat Genet. 2011.
Abstract
Through complementary application of SNP genotyping, whole-genome sequencing and imputation in 38,384 Icelanders, we have discovered a previously unidentified sick sinus syndrome susceptibility gene, MYH6, encoding the alpha heavy chain subunit of cardiac myosin. A missense variant in this gene, c.2161C>T, results in the conceptual amino acid substitution p.Arg721Trp, has an allelic frequency of 0.38% in Icelanders and associates with sick sinus syndrome with an odds ratio = 12.53 and P = 1.5 × 10⁻²⁹. We show that the lifetime risk of being diagnosed with sick sinus syndrome is around 6% for non-carriers of c.2161C>T but is approximately 50% for carriers of the c.2161C>T variant.
Figures
Figure 1
Study design and outcomes. The white boxes describe study actions performed. The blue boxes describe results from preceding actions. SSS, sick sinus syndrome.
Figure 2
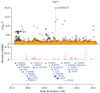
An overview of the region around c.2161C>T. The black circles show −log10 P for association with sick sinus syndrome for imputed SNPs based on whole-genome sequencing as a function of their build 36 coordinates. The orange crosses show results conditional on the effect of c.2161C>T. Neighboring genes are shown in blue. Recombination rates are reported in cM/Mb.
Figure 3
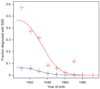
Penetrance of sick sinus syndrome among carriers and non-carriers of c.2161C>T. The red crosses represent observed penetrance of sick sinus syndrome for 10-year birth cohorts among heterozygous carriers of c.2161C>T. The red line represents the fit of the logistic model to the c.2161C>T carrier data. The blue line and crosses represent the same information for non-carriers of c.2161C>T. SSS, sick sinus syndrome.
Comment in
- Next-generation association studies for complex traits.
Zeggini E. Zeggini E. Nat Genet. 2011 Mar 29;43(4):287-8. doi: 10.1038/ng0411-287. Nat Genet. 2011. PMID: 21445070 Free PMC article.
Similar articles
- Novel mutation in the α-myosin heavy chain gene is associated with sick sinus syndrome.
Ishikawa T, Jou CJ, Nogami A, Kowase S, Arrington CB, Barnett SM, Harrell DT, Arimura T, Tsuji Y, Kimura A, Makita N. Ishikawa T, et al. Circ Arrhythm Electrophysiol. 2015 Apr;8(2):400-8. doi: 10.1161/CIRCEP.114.002534. Epub 2015 Feb 25. Circ Arrhythm Electrophysiol. 2015. PMID: 25717017 - A rare missense mutation in MYH6 associates with non-syndromic coarctation of the aorta.
Bjornsson T, Thorolfsdottir RB, Sveinbjornsson G, Sulem P, Norddahl GL, Helgadottir A, Gretarsdottir S, Magnusdottir A, Danielsen R, Sigurdsson EL, Adalsteinsdottir B, Gunnarsson SI, Jonsdottir I, Arnar DO, Helgason H, Gudbjartsson T, Gudbjartsson DF, Thorsteinsdottir U, Holm H, Stefansson K. Bjornsson T, et al. Eur Heart J. 2018 Sep 7;39(34):3243-3249. doi: 10.1093/eurheartj/ehy142. Eur Heart J. 2018. PMID: 29590334 Free PMC article. - Next-generation association studies for complex traits.
Zeggini E. Zeggini E. Nat Genet. 2011 Mar 29;43(4):287-8. doi: 10.1038/ng0411-287. Nat Genet. 2011. PMID: 21445070 Free PMC article. - Several common variants modulate heart rate, PR interval and QRS duration.
Holm H, Gudbjartsson DF, Arnar DO, Thorleifsson G, Thorgeirsson G, Stefansdottir H, Gudjonsson SA, Jonasdottir A, Mathiesen EB, Njølstad I, Nyrnes A, Wilsgaard T, Hald EM, Hveem K, Stoltenberg C, Løchen ML, Kong A, Thorsteinsdottir U, Stefansson K. Holm H, et al. Nat Genet. 2010 Feb;42(2):117-22. doi: 10.1038/ng.511. Epub 2010 Jan 10. Nat Genet. 2010. PMID: 20062063 - Structural implications of β-cardiac myosin heavy chain mutations in human disease.
Colegrave M, Peckham M. Colegrave M, et al. Anat Rec (Hoboken). 2014 Sep;297(9):1670-80. doi: 10.1002/ar.22973. Anat Rec (Hoboken). 2014. PMID: 25125180 Review.
Cited by
- Genome-wide association study of antipsychotic-induced sinus bradycardia in Chinese schizophrenia patients.
Weng S, Wang B, Li M, Chao S, Lin R, Zheng R, Yu Y, Guo S, Lin X. Weng S, et al. PLoS One. 2021 Apr 29;16(4):e0249997. doi: 10.1371/journal.pone.0249997. eCollection 2021. PLoS One. 2021. PMID: 33914752 Free PMC article. - Case Report: SCN5A mutations in three young patients with sick sinus syndrome.
Liang J, Luo S, Huang B. Liang J, et al. Front Cardiovasc Med. 2023 Dec 1;10:1294197. doi: 10.3389/fcvm.2023.1294197. eCollection 2023. Front Cardiovasc Med. 2023. PMID: 38107266 Free PMC article. - In search of low-frequency and rare variants affecting complex traits.
Panoutsopoulou K, Tachmazidou I, Zeggini E. Panoutsopoulou K, et al. Hum Mol Genet. 2013 Oct 15;22(R1):R16-21. doi: 10.1093/hmg/ddt376. Epub 2013 Aug 6. Hum Mol Genet. 2013. PMID: 23922232 Free PMC article. Review. - Atrial fibrillation: pathophysiology, genetic and epigenetic mechanisms.
Vinciguerra M, Dobrev D, Nattel S. Vinciguerra M, et al. Lancet Reg Health Eur. 2024 Feb 1;37:100785. doi: 10.1016/j.lanepe.2023.100785. eCollection 2024 Feb. Lancet Reg Health Eur. 2024. PMID: 38362554 Free PMC article. Review. - A polymorphism in IRF4 affects human pigmentation through a tyrosinase-dependent MITF/TFAP2A pathway.
Praetorius C, Grill C, Stacey SN, Metcalf AM, Gorkin DU, Robinson KC, Van Otterloo E, Kim RS, Bergsteinsdottir K, Ogmundsdottir MH, Magnusdottir E, Mishra PJ, Davis SR, Guo T, Zaidi MR, Helgason AS, Sigurdsson MI, Meltzer PS, Merlino G, Petit V, Larue L, Loftus SK, Adams DR, Sobhiafshar U, Emre NC, Pavan WJ, Cornell R, Smith AG, McCallion AS, Fisher DE, Stefansson K, Sturm RA, Steingrimsson E. Praetorius C, et al. Cell. 2013 Nov 21;155(5):1022-33. doi: 10.1016/j.cell.2013.10.022. Cell. 2013. PMID: 24267888 Free PMC article.
References
- Ferrer MI. The sick sinus syndrome in atrial disease. J. Am. Med. Assoc. 1968;206:645–646. - PubMed
- Saksena S, Camm JA. Electrophysiological Disorders of the Heart. Philadelphia: Elsevier Churchill Livingstone; 2004.
- Epstein AE, et al. ACC/AHA/HRS 2008 Guidelines for Device-Based Therapy of Cardiac Rhythm Abnormalities: a report of the American College of Cardiology/American Heart Association Task Force on Practice Guidelines (Writing Committee to Revise the ACC/AHA/NASPE 2002 Guideline Update for Implantation of Cardiac Pacemakers and Antiarrhythmia Devices) developed in collaboration with the American Association for Thoracic Surgery and Society of Thoracic Surgeons. J. Am. Coll. Cardiol. 2008;51:e1–e62. - PubMed
- Kusumoto FM, Goldschlager N. Cardiac pacing. N. Engl. J. Med. 1996;334:89–99. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 HL092217/HL/NHLBI NIH HHS/United States
- U19 HL065962/HL/NHLBI NIH HHS/United States
- HL092217/HL/NHLBI NIH HHS/United States
- U19HL065962/HL/NHLBI NIH HHS/United States
- R01 HL092217-03/HL/NHLBI NIH HHS/United States
- U19 HL065962-10/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases