A genome-wide study of DNA methylation patterns and gene expression levels in multiple human and chimpanzee tissues - PubMed (original) (raw)
A genome-wide study of DNA methylation patterns and gene expression levels in multiple human and chimpanzee tissues
Athma A Pai et al. PLoS Genet. 2011 Feb.
Abstract
The modification of DNA by methylation is an important epigenetic mechanism that affects the spatial and temporal regulation of gene expression. Methylation patterns have been described in many contexts within and across a range of species. However, the extent to which changes in methylation might underlie inter-species differences in gene regulation, in particular between humans and other primates, has not yet been studied. To this end, we studied DNA methylation patterns in livers, hearts, and kidneys from multiple humans and chimpanzees, using tissue samples for which genome-wide gene expression data were also available. Using the multi-species gene expression and methylation data for 7,723 genes, we were able to study the role of promoter DNA methylation in the evolution of gene regulation across tissues and species. We found that inter-tissue methylation patterns are often conserved between humans and chimpanzees. However, we also found a large number of gene expression differences between species that might be explained, at least in part, by corresponding differences in methylation levels. In particular, we estimate that, in the tissues we studied, inter-species differences in promoter methylation might underlie as much as 12%-18% of differences in gene expression levels between humans and chimpanzees.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
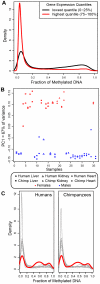
Figure 1. Genome-wide methylation patterns across all samples.
(A) Probability density functions (y-axis) of estimated promoter methylation levels (plotted as β-values on the x-axis) for genes whose expression levels are in the lowest (black) and highest (red) quartiles. (B) Principal components analysis of the methylation data from the 365 X-chromosome probes from all samples (PC1 is plotted on the y-axis, with sample indices on the x-axis). (C) Density (y-axes) histograms of β-values in humans (left) and chimpanzees (right) for 90 CpG-sites associated with 27 genes previously identified as imprinted in humans. The red lines indicate the β-value distributions in the genes with evidence for imprinting, while the grey lines are β-value distributions in ten sets of 90 randomly chosen probes.
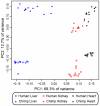
Figure 2. Principal components analysis of the methylation profiles.
This analysis only considers the 9,911 autosomal CpG sites from all samples (PC1 and PC2 are plotted on the x-axis and y-axis, respectively).
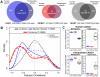
Figure 3. Conservation of tissue-specific differentially methylated regions.
(A) Venn diagrams of the number of T-DMRs classified in each species per tissue. (B) Probability density functions (y-axis) of distributions of Pearson correlations (x-axis) between methylation and gene expression levels across tissues, in human (solid lines) and chimpanzee (broken lines), for all genes expressed in at least one tissue (black), genes associated with a T-DMR in only one species (blue), and genes associated with a conserved T-DMRs (red). See Figure S4 for plots of the tissue-specific data (C) A representative example of a heart-specific T-DMR associated with the CASQ1 gene. Plotted in the left panels are the methylation β-values (y-axis), and in the right panels are the normalized gene expression levels (y-axis) in liver, kidney, and heart samples from human (top) and chimpanzee (bottom).
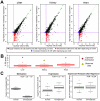
Figure 4. Inter-species methylation and gene expression differences.
(A) Scatter-plots of the p-values obtained by testing the null hypothesis of no differences in gene expression levels between human and chimpanzee before (x-axis) and after (y-axis) regressing out methylation levels. The solid purple lines correspond to a 1% FDR threshold. (B) Boxplots of the distributions, based on 1000 permutations, of the percentage of genes for which the evidence for inter-species differences in gene expression levels is expected to be reduced following the correction for methylation levels, by chance alone. Yellow points indicate the percentages seen in the actual data. (C) A representative example of the ZBTB80S gene, which is associated with inter-species promoter methylation differences in the kidney. In the left and middle panels are the human and chimpanzee methylation β-values and normalized gene expression levels, respectively. In the right panel are the normalized gene expression levels for ZBTB80S, after correcting for the methylation β-values.
Similar articles
- A comparison of gene expression and DNA methylation patterns across tissues and species.
Blake LE, Roux J, Hernando-Herraez I, Banovich NE, Perez RG, Hsiao CJ, Eres I, Cuevas C, Marques-Bonet T, Gilad Y. Blake LE, et al. Genome Res. 2020 Feb;30(2):250-262. doi: 10.1101/gr.254904.119. Epub 2020 Jan 17. Genome Res. 2020. PMID: 31953346 Free PMC article. - Divergent whole-genome methylation maps of human and chimpanzee brains reveal epigenetic basis of human regulatory evolution.
Zeng J, Konopka G, Hunt BG, Preuss TM, Geschwind D, Yi SV. Zeng J, et al. Am J Hum Genet. 2012 Sep 7;91(3):455-65. doi: 10.1016/j.ajhg.2012.07.024. Epub 2012 Aug 23. Am J Hum Genet. 2012. PMID: 22922032 Free PMC article. - Estimation of chimpanzee age based on DNA methylation.
Ito H, Udono T, Hirata S, Inoue-Murayama M. Ito H, et al. Sci Rep. 2018 Jul 3;8(1):9998. doi: 10.1038/s41598-018-28318-9. Sci Rep. 2018. PMID: 29968770 Free PMC article. - Comparative primate genomics: the year of the chimpanzee.
Ruvolo M. Ruvolo M. Curr Opin Genet Dev. 2004 Dec;14(6):650-6. doi: 10.1016/j.gde.2004.08.007. Curr Opin Genet Dev. 2004. PMID: 15531160 Review. - Differences between human and chimpanzee genomes and their implications in gene expression, protein functions and biochemical properties of the two species.
Suntsova MV, Buzdin AA. Suntsova MV, et al. BMC Genomics. 2020 Sep 10;21(Suppl 7):535. doi: 10.1186/s12864-020-06962-8. BMC Genomics. 2020. PMID: 32912141 Free PMC article. Review.
Cited by
- The relationship between regulatory changes in cis and trans and the evolution of gene expression in humans and chimpanzees.
Barr KA, Rhodes KL, Gilad Y. Barr KA, et al. Genome Biol. 2023 Sep 11;24(1):207. doi: 10.1186/s13059-023-03019-3. Genome Biol. 2023. PMID: 37697401 Free PMC article. - A cross-platform genome-wide comparison of the relationship of promoter DNA methylation to gene expression.
Plume JM, Beach SR, Brody GH, Philibert RA. Plume JM, et al. Front Genet. 2012 Feb 6;3:12. doi: 10.3389/fgene.2012.00012. eCollection 2012. Front Genet. 2012. PMID: 22363339 Free PMC article. - Evaluating Chromatin Accessibility Differences Across Multiple Primate Species Using a Joint Modeling Approach.
Edsall LE, Berrio A, Majoros WH, Swain-Lenz D, Morrow S, Shibata Y, Safi A, Wray GA, Crawford GE, Allen AS. Edsall LE, et al. Genome Biol Evol. 2019 Oct 1;11(10):3035-3053. doi: 10.1093/gbe/evz218. Genome Biol Evol. 2019. PMID: 31599933 Free PMC article. - Gene-specific DNA methylation association with serum levels of C-reactive protein in African Americans.
Sun YV, Lazarus A, Smith JA, Chuang YH, Zhao W, Turner ST, Kardia SL. Sun YV, et al. PLoS One. 2013 Aug 19;8(8):e73480. doi: 10.1371/journal.pone.0073480. eCollection 2013. PLoS One. 2013. PMID: 23977389 Free PMC article. - Insights from a chimpanzee adipose stromal cell population: opportunities for adult stem cells to expand primate functional genomics.
Pfefferle LW, Wray GA. Pfefferle LW, et al. Genome Biol Evol. 2013;5(10):1995-2005. doi: 10.1093/gbe/evt148. Genome Biol Evol. 2013. PMID: 24092797 Free PMC article.
References
- King M-C, Wilson AC. Evolution at Two Levels in Humans and Chimpanzees. Science. 1975;188:107–116. - PubMed
- Britten RJ, Davidson EH. Gene Regulation for Higher Cells: A Theory. Science. 1969:349–357. - PubMed
- Enard W, Khaitovich P, Klose J, Zöllner S, Heissig F, et al. Intra- and interspecific variation in primate gene expression patterns. Science. 2002;296:340–343. - PubMed
- Khaitovich P, Hellmann I, Enard W, Nowick K, Leinweber M, et al. Parallel patterns of evolution in the genomes and transcriptomes of humans and chimpanzees. Science. 2005;309:1850–1854. - PubMed
Publication types
MeSH terms
Grants and funding
- HHMI/Howard Hughes Medical Institute/United States
- R01 MH084703/MH/NIMH NIH HHS/United States
- GM077959/GM/NIGMS NIH HHS/United States
- GM084996/GM/NIGMS NIH HHS/United States
- MH084703/MH/NIMH NIH HHS/United States
- R01 GM077959/GM/NIGMS NIH HHS/United States
- WT_/Wellcome Trust/United Kingdom
- R01 GM084996/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases