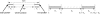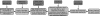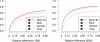Inference of isoforms from short sequence reads - PubMed (original) (raw)
Inference of isoforms from short sequence reads
Jianxing Feng et al. J Comput Biol. 2011 Mar.
Abstract
Due to alternative splicing events in eukaryotic species, the identification of mRNA isoforms (or splicing variants) is a difficult problem. Traditional experimental methods for this purpose are time consuming and cost ineffective. The emerging RNA-Seq technology provides a possible effective method to address this problem. Although the advantages of RNA-Seq over traditional methods in transcriptome analysis have been confirmed by many studies, the inference of isoforms from millions of short sequence reads (e.g., Illumina/Solexa reads) has remained computationally challenging. In this work, we propose a method to calculate the expression levels of isoforms and infer isoforms from short RNA-Seq reads using exon-intron boundary, transcription start site (TSS) and poly-A site (PAS) information. We first formulate the relationship among exons, isoforms, and single-end reads as a convex quadratic program, and then use an efficient algorithm (called IsoInfer) to search for isoforms. IsoInfer can calculate the expression levels of isoforms accurately if all the isoforms are known and infer novel isoforms from scratch. Our experimental tests on known mouse isoforms with both simulated expression levels and reads demonstrate that IsoInfer is able to calculate the expression levels of isoforms with an accuracy comparable to the state-of-the-art statistical method and a 60 times faster speed. Moreover, our tests on both simulated and real reads show that it achieves a good precision and sensitivity in inferring isoforms when given accurate exon-intron boundary, TSS, and PAS information, especially for isoforms whose expression levels are significantly high. The software is publicly available for free at http://www.cs.ucr.edu/∼jianxing/IsoInfer.html.
Figures
FIG. 1.
Expressed segments. Every exon-intron boundary introduces a boundary of some segment. Every expressed segment is a part of an exon.
FIG. 2.
(Left) A paired-end read consisting of two short reads of length _L_2 that are separated by a gap. (Right) Three consecutive intervals on an isoform.
FIG. 3.
The flow of data processing in algorithm IsoInfer.
FIG. 4.
Comparison of the accuracies of different methods in estimating isoform expression levels. The _y_-axis shows the percentage of isoforms whose estimated/calculated expression levels are within a certain relative difference range from the truth. 10 million reads (left) and 80 million reads (right) are sampled in each of the figures.
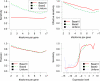
FIG. 5.
The sensitivity (top left), effective sensitivity (top right) and precision (bottom left) of IsoInfer on genes with a certain number of isoforms when different distributions of expression levels are generated. (Bottom right) Sensitivity of IsoInfer on different expression levels when different distributions of expression level are applied. In the graph, the expression levels are log_2 transformed. Expression level x corresponds to 25 · 2_x RPKM. The vertical line corresponds to expression level 1/8 = 3.125 RPKM.
FIG. 6.
The sensitivity (top left), effective sensitivity (top right) and precision (bottom left) of IsoInfer on genes with a certain number of isoforms when different combinations of type I, II, and III data are provided. (Bottom right) Sensitivity of IsoInfer on different expression levels when different combinations of type I, II, and III data are used. Again, the expression levels are log_2 transformed. Expression level x corresponds to 25 · 2_x RPKM. The vertical line corresponds to expression level 1/8 = 3.125 RPKM.
FIG. 7.
The sensitivity and precision of IsoInfer when α is set to different values.
Similar articles
- Accurate inference of isoforms from multiple sample RNA-Seq data.
Tasnim M, Ma S, Yang EW, Jiang T, Li W. Tasnim M, et al. BMC Genomics. 2015;16 Suppl 2(Suppl 2):S15. doi: 10.1186/1471-2164-16-S2-S15. Epub 2015 Jan 21. BMC Genomics. 2015. PMID: 25708199 Free PMC article. - Transcriptome assembly and isoform expression level estimation from biased RNA-Seq reads.
Li W, Jiang T. Li W, et al. Bioinformatics. 2012 Nov 15;28(22):2914-21. doi: 10.1093/bioinformatics/bts559. Epub 2012 Oct 11. Bioinformatics. 2012. PMID: 23060617 Free PMC article. - Freddie: annotation-independent detection and discovery of transcriptomic alternative splicing isoforms using long-read sequencing.
Orabi B, Xie N, McConeghy B, Dong X, Chauve C, Hach F. Orabi B, et al. Nucleic Acids Res. 2023 Jan 25;51(2):e11. doi: 10.1093/nar/gkac1112. Nucleic Acids Res. 2023. PMID: 36478271 Free PMC article. - Survey of Programs Used to Detect Alternative Splicing Isoforms from Deep Sequencing Data In Silico.
Min F, Wang S, Zhang L. Min F, et al. Biomed Res Int. 2015;2015:831352. doi: 10.1155/2015/831352. Epub 2015 Sep 3. Biomed Res Int. 2015. PMID: 26421304 Free PMC article. Review. - From words to complete phrases: insight into single-cell isoforms using short and long reads.
Joglekar A, Foord C, Jarroux J, Pollard S, Tilgner HU. Joglekar A, et al. Transcription. 2023 Jun-Oct;14(3-5):92-104. doi: 10.1080/21541264.2023.2213514. Epub 2023 Jun 14. Transcription. 2023. PMID: 37314295 Free PMC article. Review.
Cited by
- Protocol for transcriptome assembly by the TransBorrow algorithm.
Zhao D, Liu J, Yu T. Zhao D, et al. Biol Methods Protoc. 2023 Nov 1;8(1):bpad028. doi: 10.1093/biomethods/bpad028. eCollection 2023. Biol Methods Protoc. 2023. PMID: 38023349 Free PMC article. Review. - StringFix: an annotation-guided transcriptome assembler improves the recovery of amino acid sequences from RNA-Seq reads.
Lee J, Kim M, Han K, Yoon S. Lee J, et al. Genes Genomics. 2023 Dec;45(12):1599-1609. doi: 10.1007/s13258-023-01458-7. Epub 2023 Oct 14. Genes Genomics. 2023. PMID: 37837515 - Counting pseudoalignments to novel splicing events.
Borozan L, Rojas Ringeling F, Kao SY, Nikonova E, Monteagudo-Mesas P, Matijević D, Spletter ML, Canzar S. Borozan L, et al. Bioinformatics. 2023 Jul 1;39(7):btad419. doi: 10.1093/bioinformatics/btad419. Bioinformatics. 2023. PMID: 37432342 Free PMC article. - Rare Does Not Mean Worthless: How Rare Diseases Have Shaped Neurodevelopment Research in the NGS Era.
Zaghi M, Banfi F, Bellini E, Sessa A. Zaghi M, et al. Biomolecules. 2021 Nov 17;11(11):1713. doi: 10.3390/biom11111713. Biomolecules. 2021. PMID: 34827709 Free PMC article. Review. - TransBorrow: genome-guided transcriptome assembly by borrowing assemblies from different assemblers.
Yu T, Mu Z, Fang Z, Liu X, Gao X, Liu J. Yu T, et al. Genome Res. 2020 Aug;30(8):1181-1190. doi: 10.1101/gr.257766.119. Epub 2020 Aug 17. Genome Res. 2020. PMID: 32817072 Free PMC article.
References
- Bertone P. Stolc V. Royce T.E., et al. Global identification of human transcribed sequences with genome tiling arrays. Science. 2004;306:2242–2246. - PubMed
- Bishop C.M. Pattern Recognition and Machine Learning. Springer; New York: 2007.
- Boguski M.S. The turning point in genome research. Trends Biochem. Sci. 1995;20:295–296. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 2R01LM008991/LM/NLM NIH HHS/United States
- R01 AI078885-01A2/AI/NIAID NIH HHS/United States
- R01 LM008991/LM/NLM NIH HHS/United States
- AI078885/AI/NIAID NIH HHS/United States
- R01 AI078885/AI/NIAID NIH HHS/United States
- R01 LM008991-04/LM/NLM NIH HHS/United States
LinkOut - more resources
Full Text Sources