Disruption of a conserved region of Xist exon 1 impairs Xist RNA localisation and X-linked gene silencing during random and imprinted X chromosome inactivation - PubMed (original) (raw)
Disruption of a conserved region of Xist exon 1 impairs Xist RNA localisation and X-linked gene silencing during random and imprinted X chromosome inactivation
Claire E Senner et al. Development. 2011 Apr.
Abstract
In XX female mammals a single X chromosome is inactivated early in embryonic development, a process that is required to equalise X-linked gene dosage relative to XY males. X inactivation is regulated by a cis-acting master switch, the Xist locus, the product of which is a large non-coding RNA that coats the chromosome from which it is transcribed, triggering recruitment of chromatin modifying factors that establish and maintain gene silencing chromosome wide. Chromosome coating and Xist RNA-mediated silencing remain poorly understood, both at the level of RNA sequence determinants and interacting factors. Here, we describe analysis of a novel targeted mutation, Xist(INV), designed to test the function of a conserved region located in exon 1 of Xist RNA during X inactivation in mouse. We show that Xist(INV) is a strong hypomorphic allele that is appropriately regulated but compromised in its ability to silence X-linked loci in cis. Inheritance of Xist(INV) on the paternal X chromosome results in embryonic lethality due to failure of imprinted X inactivation in extra-embryonic lineages. Female embryos inheriting Xist(INV) on the maternal X chromosome undergo extreme secondary non-random X inactivation, eliminating the majority of cells that express the Xist(INV) allele. Analysis of cells that express Xist(INV) RNA demonstrates reduced association of the mutant RNA to the X chromosome, suggesting that conserved sequences in the inverted region are important for Xist RNA localisation.
Figures
Fig. 1.
Strategy for generating a targeted inversion of Xist exon 1 conserved sequences. (A) Dotplot analysis of mouse and human Xist/XIST cDNA. Full length mouse and human Xist/XIST cDNA sequences were compared by dotplot analysis using the EMBOSS dotmatcher program (Rice et al., 2000). Two sets of parameters with different stringencies were used to demonstrate overall homology between two sequences (window size 50, threshold 45) or to highlight longer stretches with high homology (window size 150, threshold 95). Exon structure for each transcript is shown above (mouse) or alongside (human) the dotplots. Mouse intron 7, which contributes to a proportion of splice variant transcripts, is shown as a white box. Exon 4, the most conserved region, is shown in blue on the schematic and encircled in blue on the dotplot. The region of inversion is indicated with red bracketed arrows and encircled in red on the dotplot. Note the extended homology over the whole region of the inversion. (B) A schematic representation of the previously targeted Xist allele with a deletion of exon 4 (Xist_Δ_ex4) (Caparros et al., 2002) and the pNBXT1 targeting construct. Conserved repetitive elements A-F within exons 1 and 7 are shown as shaded boxes. LoxP sequences (grey triangles) flank a Pgk promoter-driven neomycin (neo) selection cassette (pale grey rectangle) between 5′ and 3′ arms of homology (dark grey rectangles). The positions of the arms of homology are indicated with black lines below the schematic. A diptheria-toxin A negative selection cassette (white rectangle) was included in the targeting construct. (C) The resulting homologous recombinant allele (Xistneo) carrying both a deletion of Xist exon 4 (replaced by a LoxP sequence) and a floxed neo cassette in reverse orientation. (D) Transient expression of Cre-recombinase generates a deletion of the neo cassette (Xist_Δ_neo) and/or inversion (XistINV) of the region between exon 1 and intron 4 (5947 bp to 13,670 bp downstream of the transcriptional start site).
Fig. 2.
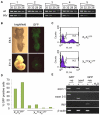
Heterozygous female embryos with a maternally transmitted XistINV allele exhibit secondary non-random X inactivation. (A) Allele-specific RT-PCR analysis of Xist from X INVmXp adult female kidneys (females 1-5). Top panel (wt) shows amplification with wild-type Xist allele specific primers (e and l) and the bottom panel presents amplification with XistINV specific primers (b and e). Numbers above the top panel show the number of cycles used. Note the six to nine cycle difference in amplification of wild-type versus XistINV fragments between different females. (B) Light and GFP microscopy images of E8.5 and E11.5 embryos with a maternally inherited XistINV allele and a paternally inherited GFP transgene. The diminishing proportion of green cells indicates secondary non-random X inactivation. (C) Examples of FACS histograms showing relative proportion of GFP-negative (M1) and GFP-positive (M2) cells in XmXp GFP and Xm INVXp GFP primary fibroblast cell lines established from E14.5 females. The _x_-axis represents the logarithmic level of GFP fluorescence and the _y_-axis shows the number of cells. 50,000 total events were acquired for each sample. (D) Quantitative analysis of FACS data obtained for two primary control fibroblast cultures (XmX GFPp) and for four primary fibroblast cultures from mutant heterozygous E14.5 embryos (Xm INVXp GFP). (E) RT-PCR analysis of the Xist transcripts produced by fibroblast cell lines expressing wild-type Xist, Xist_Δ_ex4 or XistINV. Top panel, primers amplifying a sequence upstream of the inversion in exon 1 (a and h); second panel, primers situated in exon 3 and exon 5, respectively (e and m); third panel, primers flanking the inversion break-point that only amplify from the XistINV allele (b and e); bottom panel, β-actin loading control. + and − indicate reactions set up with and without reverse transcriptase, respectively. Positions of all Xist primers used are shown in Fig. S1 in the supplementary material.
Fig. 3.
Embryo lethality in XX embryos inheriting XistINV on the paternal X chromosome. Embryos were dissected from uteri at E6.5-10.5. Xm carries the wild-type Xist allele and so male (XmY) embryos are wild type. Xp carries the XistINV allele and so females are heterozygous for the inversion. The embryos shown are representatives from three litters dissected at each gestational age. Xm, maternally transmitted X chromosome; Xp, paternally transmitted X chromosome. Scale bars: 500 μm.
Fig. 4.
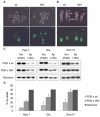
Failure of X-linked gene silencing in female embryos carrying a paternally inherited XistINV allele. (A,B) Light and GFP microscopy images of E3.5 (A) and E6.5 (B) female embryos with a paternally inherited XistINV allele and XGFP transgene. GFP expression in blastocysts (E3.5) and extra-embryonic tissue at E6.5 indicates that XistINV cannot establish or maintain appropriate levels of X-linked gene silencing. The arrowhead marks a single blastocyst with a maternally inherited GFP transgene to show the intensity of full activation of the transgene. Scale bars: 100 μm in A, 500 μm in B. emb, embryonic part; ex, extra-embryonic part; wt, wild type. (C) Allele-specific gene expression analysis of three X-linked genes: Pgk-1, Gla and Smc1l1. E3.5 embryos were obtained from crosses between either wild-type or _XistINV_-carrying males and PGK females. Polymorphisms arising between the PGK strain and the targeted 129 strain allowed expression from Xm (PGK) and Xp (129) alleles to be distinguished by single nucleotide primer extension (SNuPE). Female embryos were identified by GFP expression and pooled together for analysis. At least six embryos were included in each pool and at least four pools were analysed. Examples show duplicate loading of RT-PCR reactions detecting the presence of expression from Xp (129) and Xm (PGK) alleles. PGK × 129 F1 genomic DNA was also included in the analysis as a control. (D) The percentage of total expression coming from the Xp allele in wild-type and _XistINV_-carrying embryos. PGK × 129 F1 genomic PCR fragments were used to normalise the data. Xp, paternally inherited X chromosome; Xm, maternally inherited X chromosome. Error bars indicate s.d.
Fig. 5.
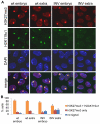
Reduced H3K27me3 and H2AK119u1 on the Xi in female embryos with a paternally inherited XistINV allele. (A) Immunofluorescence images taken of cells from wild-type and XistINV female embryonic and extra-embryonic parts at E6.5 stained for H3K27me3 (red) and H2AK119u1 (green). DNA was counterstained with DAPI (blue). (B) Scoring data showing the numbers of H3K27me3 and H2AK119u1 domains detected by immunofluorescence in cells from wild-type and XistINV female embryonic and extra-embryonic parts at E6.5. Error bars indicate s.d.
Fig. 6.
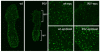
Reduced Xist domains in extra-embryonic tissue of female embryos with a paternally inherited XistINV allele. RNA FISH on female embryos collected at E6.5 reveals Xist RNA (green) coating of the inactive X chromosome throughout the whole of the wild-type female embryo (left panel) and in the increased magnifications of the epiblast (epi) and ectoplacental cone (epc) (middle right panels). Xist RNA coating can be seen at levels comparable to wild type in the XistINV epiblast only (middle left and bottom right panels). In the extra-embryonic part of the embryo the domains appear smaller and weaker (middle left and top right panels).
Fig. 7.
XistINV RNA does not localise efficiently to the X chromosome. Representative examples illustrating Xist RNA localisation (green) determined by RNA FISH on (A) interphase and (B) metaphase chromosomes from fibroblast cell lines expressing wild-type Xist, Xist_Δ_ex4 or XistINV. DNA was counterstained with DAPI (blue).
Similar articles
- Incomplete X-inactivation initiated by a hypomorphic Xist allele in the mouse.
Hoki Y, Ikeda R, Mise N, Sakata Y, Ohhata T, Sasaki H, Abe K, Sado T. Hoki Y, et al. Development. 2011 Jul;138(13):2649-59. doi: 10.1242/dev.061226. Epub 2011 May 25. Development. 2011. PMID: 21613321 - Evidence of Xist RNA-independent initiation of mouse imprinted X-chromosome inactivation.
Kalantry S, Purushothaman S, Bowen RB, Starmer J, Magnuson T. Kalantry S, et al. Nature. 2009 Jul 30;460(7255):647-51. doi: 10.1038/nature08161. Epub 2009 Jul 1. Nature. 2009. PMID: 19571810 Free PMC article. - Genes flanking Xist in mouse and human are separated on the X chromosome in American marsupials.
Shevchenko AI, Zakharova IS, Elisaphenko EA, Kolesnikov NN, Whitehead S, Bird C, Ross M, Weidman JR, Jirtle RL, Karamysheva TV, Rubtsov NB, VandeBerg JL, Mazurok NA, Nesterova TB, Brockdorff N, Zakian SM. Shevchenko AI, et al. Chromosome Res. 2007;15(2):127-36. doi: 10.1007/s10577-006-1115-9. Epub 2007 Mar 5. Chromosome Res. 2007. PMID: 17333537 Free PMC article. - Advances in understanding chromosome silencing by the long non-coding RNA Xist.
Sado T, Brockdorff N. Sado T, et al. Philos Trans R Soc Lond B Biol Sci. 2013 Jan 5;368(1609):20110325. doi: 10.1098/rstb.2011.0325. Philos Trans R Soc Lond B Biol Sci. 2013. PMID: 23166390 Free PMC article. Review. - What makes the maternal X chromosome resistant to undergoing imprinted X inactivation?
Sado T. Sado T. Philos Trans R Soc Lond B Biol Sci. 2017 Nov 5;372(1733):20160365. doi: 10.1098/rstb.2016.0365. Philos Trans R Soc Lond B Biol Sci. 2017. PMID: 28947661 Free PMC article. Review.
Cited by
- RNA-sequencing from single nuclei.
Grindberg RV, Yee-Greenbaum JL, McConnell MJ, Novotny M, O'Shaughnessy AL, Lambert GM, Araúzo-Bravo MJ, Lee J, Fishman M, Robbins GE, Lin X, Venepally P, Badger JH, Galbraith DW, Gage FH, Lasken RS. Grindberg RV, et al. Proc Natl Acad Sci U S A. 2013 Dec 3;110(49):19802-7. doi: 10.1073/pnas.1319700110. Epub 2013 Nov 18. Proc Natl Acad Sci U S A. 2013. PMID: 24248345 Free PMC article. - Pan-cancer analysis of LncRNA XIST and its potential mechanisms in human cancers.
Han W, Shi CT, Ma J, Chen H, Shao QX, Gao XJ, Zhou Y, Gu JF, Wang HN. Han W, et al. Heliyon. 2022 Sep 28;8(10):e10786. doi: 10.1016/j.heliyon.2022.e10786. eCollection 2022 Oct. Heliyon. 2022. PMID: 36212008 Free PMC article. - In-cell identification and measurement of RNA-protein interactions.
Graindorge A, Pinheiro I, Nawrocka A, Mallory AC, Tsvetkov P, Gil N, Carolis C, Buchholz F, Ulitsky I, Heard E, Taipale M, Shkumatava A. Graindorge A, et al. Nat Commun. 2019 Nov 22;10(1):5317. doi: 10.1038/s41467-019-13235-w. Nat Commun. 2019. PMID: 31757954 Free PMC article. - Functional impacts of non-coding RNA processing on enhancer activity and target gene expression.
Ntini E, Marsico A. Ntini E, et al. J Mol Cell Biol. 2019 Oct 25;11(10):868-879. doi: 10.1093/jmcb/mjz047. J Mol Cell Biol. 2019. PMID: 31169884 Free PMC article. Review. - The Xist lncRNA exploits three-dimensional genome architecture to spread across the X chromosome.
Engreitz JM, Pandya-Jones A, McDonel P, Shishkin A, Sirokman K, Surka C, Kadri S, Xing J, Goren A, Lander ES, Plath K, Guttman M. Engreitz JM, et al. Science. 2013 Aug 16;341(6147):1237973. doi: 10.1126/science.1237973. Epub 2013 Jul 4. Science. 2013. PMID: 23828888 Free PMC article.
References
- Barakat T. S., Jonkers I., Monkhorst K., Gribnau J. (2010). X-changing information on X inactivation. Exp. Cell Res. 316, 679-687 - PubMed
- Belyaev N., Keohane A. M., Turner B. M. (1996). Differential underacetylation of histones H2A, H3 and H4 on the inactive X chromosome in human female cells. Hum. Genet. 97, 73-78 - PubMed
- Borsani G., Tonlorenzi R., Simmler M. C., Dandolo L., Arnaud D., Capra V., Grompe M., Pizzuti A., Muzny D., Lawrence C., et al. (1991). Characterization of a murine gene expressed from the inactive X chromosome. Nature 351, 325-329 - PubMed
- Brockdorff N., Ashworth A., Kay G. F., Cooper P., Smith S., McCabe V. M., Norris D. P., Penny G. D., Patel D., Rastan S. (1991). Conservation of position and exclusive expression of mouse Xist from the inactive X chromosome. Nature 351, 329-331 - PubMed
- Brockdorff N., Ashworth A., Kay G. F., McCabe V. M., Norris D. P., Cooper P. J., Swift S., Rastan S. (1992). The product of the mouse Xist gene is a 15kb inactive X-specific transcript containing no conserved ORF and located in the nucleus. Cell 71, 515-526 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases