The ER stress factor XBP1s prevents amyloid-beta neurotoxicity - PubMed (original) (raw)
. 2011 Jun 1;20(11):2144-60.
doi: 10.1093/hmg/ddr100. Epub 2011 Mar 9.
Affiliations
- PMID: 21389082
- PMCID: PMC3090193
- DOI: 10.1093/hmg/ddr100
The ER stress factor XBP1s prevents amyloid-beta neurotoxicity
Sergio Casas-Tinto et al. Hum Mol Genet. 2011.
Abstract
Alzheimer's disease (AD) is an incurable neurodegenerative disorder clinically characterized by progressive cognitive impairment. A prominent pathologic hallmark in the AD brain is the abnormal accumulation of the amyloid-β 1-42 peptide (Aβ), but the exact pathways mediating Aβ neurotoxicity remain enigmatic. Endoplasmic reticulum (ER) stress is induced during AD, and has been indirectly implicated as a mediator of Aβ neurotoxicity. We report here that Aβ activates the ER stress response factor X-box binding protein 1 (XBP1) in transgenic flies and in mammalian cultured neurons, yielding its active form, the transcription factor XBP1s. XBP1s shows neuroprotective activity in two different AD models, flies expressing Aβ and mammalian cultured neurons treated with Aβ oligomers. Trying to identify the mechanisms mediating XBP1s neuroprotection, we found that in PC12 cells treated with Aβ oligomers, XBP1s prevents the accumulation of free calcium (Ca(2+)) in the cytosol. This protective activity can be mediated by the downregulation of a specific isoform of the ryanodine Ca(2+) channel, RyR3. In support of this observation, a mutation in the only ryanodine receptor (RyR) in flies also suppresses Aβ neurotoxicity, indicating the conserved mechanisms between the two AD models. These results underscore the functional relevance of XBP1s in Aβ toxicity, and uncover the potential of XBP1 and RyR as targets for AD therapeutics.
Figures
Figure 1.
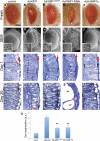
XBP1 suppresses Aβ neurotoxicity in the fly eye. Fresh eyes (A–E), scanning electron micrographs (F–J) and frontal eye sections at day 1 (K–O) and day 20 (P–T) of control flies (gmr-Gal4/UAS–GFP; A, F, K, P), flies expressing Aβ (gmr-Gal4/UAS–Aβ/UAS–GFP; B, G, L, Q) or Aβ in combination with Drosophila XBP1 (gmr-Gal4/UAS–Aβ/XBP1d08698; C, H, M, R), dXBP1–RNAi (gmr-Gal4/UAS–Aβ/UAS–XBP1–RNAi; D, I, N, S) or mXBP1s (gmr-Gal4/UAS–Aβ/UAS–mXBP1s; E, J, O, T). Compared with control flies (A and F), the eyes of flies expressing Aβ alone are small, highly disorganized and contain black, necrotic spots (B and G). The retina at day 1 is thinner (from arrow to dotted line), the photoreceptors (boxes) are very disorganized and the lenses are poorly differentiated (arrows) (K and L). By day 20, the retinas of the Aβ flies are more disorganized and vacuolated (P and Q, arrowheads). The eyes of flies co-expressing Aβ and dXBP1 (C and H) or mXBP1s (E and J) are bigger, better organized and do not contain necrotic spots. At day 1, the retinas of these flies are deeper (arrow to dotted line), the photoreceptors are better differentiated (boxes) and the lenses (arrows) are partially rescued (M and O). By day 20, these retinas maintain their organization with clearly visible photoreceptors (R and T, boxes). Flies co-expressing dXBP1–RNAi exhibit very small, disorganized and depigmented eyes (D and I) and their retinas are very thin with poorly developed lenses (N, arrow) and photoreceptors (N, box). By day 20, the retinas are mostly vacuolated (arrowheads) and no photoreceptors are visible (S). (U) Quantitation of eye phenotypes (1 = normal, 5 = small, disorganized eyes) shows that both XBP1d08698 and mXBP1s significantly reduced Aβ neurotoxicity (_n_= 4, P< 0.01).
Figure 2.
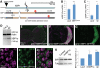
Unconventional splicing of XBP1. (A) Genomic map of the XBP1 locus in Drosophila. The XBP1d08698 insertion that regulates XBP1 expression is located in the 5′ UTR. XBP1 produces two isoforms that differ in the inclusion (XBP1u) or exclusion (XBP1s) of a 23-nucleotide intron subjected to cytoplasmic splicing. The site for _Pst_I digestion inside the 23 bp intron is also shown. (B) The XBP1d08698 insertion induces XBP1 expression. Combination of da-Gal4 with XBP1d08698 induces a 40% increase in total XBP1 mRNA compared with flies expressing LacZ (control) by qPCR. (C) Aβ induces XBP1 expression. Flies expressing Aβ under the control of da-Gal4 induce a 3.5-fold increase in total XBP1 transcripts compared with LacZ control flies. In (C) and (D), XBP1 expression was normalized to Actin. (D) Aβ induces XBP1 unconventional splicing. Flies expressing Aβ under the control of da-Gal4 accumulate higher amounts of the XBP1s isoform than control flies expressing LacZ. Actin is shown as loading control. (E–J) Aβ activates XBP1 splicing in vivo. (E) Control flies expressing the XBP1–GFP sensor in the eye (gmr-Gal4/UAS–XBP1–GFP) do not produce GFP. (F and G) Flies co-expressing Aβ and the sensor (gmr-Gal4/UAS–Aβ/UAS–XBP1–GFP) accumulate Aβ (magenta, F) and high levels of GFP (green, F and G) in the same territory of the developing eye. (H–J) All the cells expressing Aβ also accumulate GFP, although the two signals do not co-localize because Aβ is in the membrane and GFP is nuclear. (K) Both mXBP1s and Aβ induce Grp78/BiP upregulation. Heads from flies expressing a control transgene (LacZ), mXBP1s or Aβ were homogenized, resolved in western blot and incubated with BiP and Tubulin antibodies. BiP appears significantly upregulated in flies expressing mXBP1s and Aβ (_n_= 3).
Figure 3.
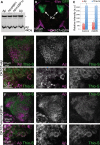
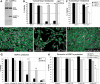
XBP1 does not affect Aβ accumulation in brain neurons. (A) Western blot shows total Aβ accumulation in Drosophila brains expressing Aβ alone (lanes 1 and 4) or co-expressing dXBP1 (lane 2) or mXBP1s (lane 3). Levels of total Aβ are the same in the three conditions. *Indicates an unspecific band. (B) Z-axis projection of a 20 µm optical section of the posterior brain of Drosophila showing accumulation of GFP in the Kenyon cells (arrows, OK107-Gal4/CD8–GFP). The cortex was labeled with anti-Elav (magenta) to include the outline of the brain. (C) Quantification of fluorescent signal for Aβ and thioflavine-S in (D) and (F). The co-expression of mXBP1s did not significantly change the levels of Aβ (red bars) or thioflavine-S (blue bars). (D and F) Detail of the Kenyon cells showing total Aβ expression (magenta) and thioflavine-S fluorescence (green) in flies only expressing Aβ (D) or co-expressing mXBP1s (F). (E and G) At higher magnification, neither the distribution of Aβ (magenta) nor the intensity of thioflavine-S fluorescence (green) is affected by mXBP1 co-expression.
Figure 4.
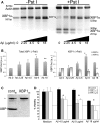
Aβ induces accumulation of neuroprotective XBP1s in mammalian neurons. (A) Accumulation of XBP1 transcripts in PC12 cells treated for 6h with a gradient of Aβ oligomers. Half of the RT–PCR reaction was digested with _Pst_I to cleave the XBP1u transcripts (right). Actin was also amplified as internal loading control. Untreated cells (0) accumulate very low levels of XBP1s, but in the presence of Aβ, the _Pst_I-resistant XBP1s accumulates in a dose-dependent manner. (B) Quantitation of total XBP1, XBP1s and XBP1u from two independent gels. The levels of total XBP1 increase with the amount of Aβ, with a maximum corresponding to 9 µg/ml. The levels of XBP1s parallel the activation curve of total XBP1, while XBP1u levels decrease with increasing amounts of Aβ. (C and D) Endogenous XBP1 protects against Aβ cytotoxicity. (C) A siRNA against XBP1 (XBP1SI) eliminates the XBP1 protein in PC12 cell extracts by western blot, while a control siRNA (contSI) does not change XBP1 levels. An unspecific band (*) is detected with the anti-XBP1 antibody. (D) The dose-dependent toxicity of Aβ oligomers in WT cells is shown in black bars. Cell viability is compromised at 18 µg/ml in 6 h treatments. Silencing of XBP1 transcripts with two independent siRNAs (XBP1SI–1 [grey] and XBP1SI–2 [white]) results in significantly reduced cell viability in the presence of Aβ oligomers (_n_= 3), even at subtoxic Aβ treatments (4.5 and 9 µg/ml). *P< 0.05, **P< 0.01.
Figure 5.
XBP1s protects against Aβ cytotoxicity. (A) Detection of mXBP1s mRNA in PC12 cells stably transfected with CMV-mXBP1s using specific primers for mouse XBP1. (B and C) mXBP1s overexpression confers resistance to tunicamycin. mXBP1s cells (grey) survive tunicamycin concentrations that kill control cells (black) (B). The protective activity of mXBP1s against tunicamycin is reverted by pre-treatment with siRNA against XBP1 (C). (D–H) mXBP1s prevents cytotoxicity of Aβ oligomers. Representative fields show the effects of Aβ on cell viability: non-treated control (WT) cells (D), and control cells (E) and mXBP1s cells (F) treated with Aβ oligomers at 18 µg/ml. Cells are stained with anti-Tubulin (green) and DAPI (magenta). (G) Aβ causes a dramatic drop in viability at 18 µg/ml in control cells (black) after an 8 h incubation. However, mXBP1s (grey) completely protects PC12 neurons treated with Aβ at 18 µg/ml, and partially protects at higher concentrations. (H) The protective activity of mXBP1s (black bars) is reverted by pre-treatment with XBP1SI (white), but not by a control RNAi (grey). *P< 0.05, **P< 0.01, ***P< 0.001.
Figure 6.
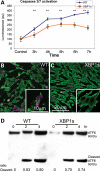
XBP1s prevents Aβ-dependent caspase activation, but not ER stress. (A) Temporal analysis of caspase-3 and -7 activation in WT and mXBP1s cells treated with Aβ oligomers at 18 µg/ml. (A) WT cells (blue diamonds) progressively accumulate activated caspases over the course of the experiment. In mXBP1s cells (orange squares), activated caspase levels peak at 5 h, then decrease in the last 2h (_n_= 3, **P< 0.01). (B and C) Distribution of activated (act)-caspase-3 in PC12 cells treated with oligomeric Aβ at 18 µg/ml. WT cells appear small and round, and accumulate act-caspase-3 (B), whereas mXBP1s cells preserve their morphology and accumulate very little act-caspase-3 (C). (D) Western blot shows ATF6 activation in WT and mXBP1s cells treated with a subtoxic dose of Aβ oligomers (9 µg/ml). After a 4 h incubation, both WT (0.83 ± 0.0075 and 0.8 ± 0.03) and mXBP1s cells (0.7 ± 0.026 and 0.74 ± 0.023) accumulate cleaved ATF6. After a 4 h incubation, both WT and mXBP1s cells accumulate cleaved ATF6. There is no significant difference in the ratio of cleaved ATF6 between WT and mXBP1s, although mXBP1s accumulated less-activated ATF6 in two different time points.
Figure 7.
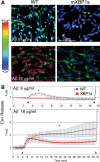
XBP1s prevents Aβ-dependent calcium release. (A) Representative micrographs of luminal Ca2+ release measured by fura2 in PC12 cells treated with oligomeric Aβ. (B) Longitudinal analysis of Ca2+ release from the same samples. A subtoxic concentration of Aβ (9 µg/ml) elicits moderate Ca2+ release into the cytosol in WT cells (blue) that are reversible after a few minutes. On the other hand, mXBP1s cells (red) do not release Ca2+ at this concentration of Aβ. At toxic concentrations (18 µg/ml), Aβ oligomers induce high levels of cytosolic Ca2+ in normal cells that continue to rise until the end of the experiment. Fifty percent of mXBP1s cells also release Ca2+ early on, although at moderate levels, and this effect is reverted later on. In (B), the first arrowhead indicates the addition of Aβ at the start of the experiment. The second arrowhead indicates the addition of ionomycin to terminate the experiment by liberating all intraluminal Ca2+ stores. Two-way ANOVA shows significant differences between the WT and mXBP1s (**P< 0.05).
Figure 8.
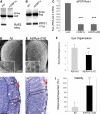
Reduced ryanodine calcium channels mediate XBP1 neuroprotection. (A) Expression of RyR3 is severely downregulated by XBP1s in PC12 cells. The RyR3 isoform is expressed in PC12 cells, but the RyR3 transcripts are almost undetectable in cells expressing mXBP1s. Actin was amplified as loading control. (B) WFS1, a known target of XBP1s, is upregulated 1.5-fold in cells expressing mXBP1s. (C) Analysis of Drosophila Rya-r transcripts by qPCR indicates that mXBP1s expression reduces Rya-r levels by 80% even in flies also expressing Aβ (_n_= 8, P< 0.001). (D–H) Reduced Rya-r activity rescues the eye degeneration induced by Aβ. Flies expressing Aβ display disorganized ommatidia (D) and small photoreceptors (G, box). In contrast, flies also carrying the null allele Rya-r16 have better-organized ommatidia (E), and deeper retinas, better differentiated lenses (arrow) and elongated photoreceptors (box) (H). (F) The distribution of eye phenotypes (1 = normal, 5 = small, disorganized eyes) shows a significant rescue of eye morphology in flies also carrying the Rya-r deletion (_n_= 4, P< 0.001). (I) Reduced Rya-r activity rescues the lethality induced by Aβ. Expression of Aβ reduces adult eclosion to 20% with respect to control siblings (grey, _n_= 6). Flies that also carrying the null allele Rya-r16 present normal viability (black, _n_= 3, P< 0.01). In (F) and (I), n is the number of vials analyzed, each producing at least 50 flies.
Similar articles
- Ryanodine receptor blockade reduces amyloid-β load and memory impairments in Tg2576 mouse model of Alzheimer disease.
Oulès B, Del Prete D, Greco B, Zhang X, Lauritzen I, Sevalle J, Moreno S, Paterlini-Bréchot P, Trebak M, Checler F, Benfenati F, Chami M. Oulès B, et al. J Neurosci. 2012 Aug 22;32(34):11820-34. doi: 10.1523/JNEUROSCI.0875-12.2012. J Neurosci. 2012. PMID: 22915123 Free PMC article. - An endoplasmic-reticulum-specific apoptotic pathway is involved in prion and amyloid-beta peptides neurotoxicity.
Ferreiro E, Resende R, Costa R, Oliveira CR, Pereira CM. Ferreiro E, et al. Neurobiol Dis. 2006 Sep;23(3):669-78. doi: 10.1016/j.nbd.2006.05.011. Epub 2006 Jul 17. Neurobiol Dis. 2006. PMID: 16844381 - Amyloid-beta-(1-42) increases ryanodine receptor-3 expression and function in neurons of TgCRND8 mice.
Supnet C, Grant J, Kong H, Westaway D, Mayne M. Supnet C, et al. J Biol Chem. 2006 Dec 15;281(50):38440-7. doi: 10.1074/jbc.M606736200. Epub 2006 Oct 18. J Biol Chem. 2006. PMID: 17050533 - Alterations of the Endoplasmic Reticulum (ER) Calcium Signaling Molecular Components in Alzheimer's Disease.
Chami M, Checler F. Chami M, et al. Cells. 2020 Dec 1;9(12):2577. doi: 10.3390/cells9122577. Cells. 2020. PMID: 33271984 Free PMC article. Review. - Amyloid beta-peptide (1-42)-induced oxidative stress and neurotoxicity: implications for neurodegeneration in Alzheimer's disease brain. A review.
Butterfield DA. Butterfield DA. Free Radic Res. 2002 Dec;36(12):1307-13. doi: 10.1080/1071576021000049890. Free Radic Res. 2002. PMID: 12607822 Review.
Cited by
- Purification of transcripts and metabolites from Drosophila heads.
Jensen K, Sanchez-Garcia J, Williams C, Khare S, Mathur K, Graze RM, Hahn DA, McIntyre LM, Rincon-Limas DE, Fernandez-Funez P. Jensen K, et al. J Vis Exp. 2013 Mar 15;(73):e50245. doi: 10.3791/50245. J Vis Exp. 2013. PMID: 23524378 Free PMC article. - Drosophila models of proteinopathies: the little fly that could.
Rincon-Limas DE, Jensen K, Fernandez-Funez P. Rincon-Limas DE, et al. Curr Pharm Des. 2012;18(8):1108-22. doi: 10.2174/138161212799315894. Curr Pharm Des. 2012. PMID: 22288402 Free PMC article. Review. - Intrinsic determinants of prion protein neurotoxicity in Drosophila: from sequence to (dys)function.
Cembran A, Fernandez-Funez P. Cembran A, et al. Front Mol Neurosci. 2023 Aug 14;16:1231079. doi: 10.3389/fnmol.2023.1231079. eCollection 2023. Front Mol Neurosci. 2023. PMID: 37645703 Free PMC article. Review. - Novel neuroprotective function of apical-basal polarity gene crumbs in amyloid beta 42 (aβ42) mediated neurodegeneration.
Steffensmeier AM, Tare M, Puli OR, Modi R, Nainaparampil J, Kango-Singh M, Singh A. Steffensmeier AM, et al. PLoS One. 2013 Nov 18;8(11):e78717. doi: 10.1371/journal.pone.0078717. eCollection 2013. PLoS One. 2013. PMID: 24260128 Free PMC article. - Endoplasmic Reticulum Stress-Associated Neuronal Death and Innate Immune Response in Neurological Diseases.
Shi M, Chai Y, Zhang J, Chen X. Shi M, et al. Front Immunol. 2022 Jan 10;12:794580. doi: 10.3389/fimmu.2021.794580. eCollection 2021. Front Immunol. 2022. PMID: 35082783 Free PMC article. Review.
References
- Haass C., Selkoe D.J. Soluble protein oligomers in neurodegeneration: lessons from the Alzheimer's amyloid beta-peptide. Nat. Rev. Mol. Cell Biol. 2007;8:101–112. - PubMed
- Hardy J. The amyloid hypothesis for Alzheimer's disease: a critical reappraisal. J. Neurochem. 2009;110:1129–1134. - PubMed
- Oddo S., Caccamo A., Kitazawa M., Tseng B.P., LaFerla F.M. Amyloid deposition precedes tangle formation in a triple transgenic model of Alzheimer's disease. Neurobiol. Aging. 2003;24:1063–1070. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous