CREBBP mutations in relapsed acute lymphoblastic leukaemia - PubMed (original) (raw)
. 2011 Mar 10;471(7337):235-9.
doi: 10.1038/nature09727.
Jinghui Zhang, Lawryn H Kasper, Stephanie Lerach, Debbie Payne-Turner, Letha A Phillips, Sue L Heatley, Linda Holmfeldt, J Racquel Collins-Underwood, Jing Ma, Kenneth H Buetow, Ching-Hon Pui, Sharyn D Baker, Paul K Brindle, James R Downing
Affiliations
- PMID: 21390130
- PMCID: PMC3076610
- DOI: 10.1038/nature09727
CREBBP mutations in relapsed acute lymphoblastic leukaemia
Charles G Mullighan et al. Nature. 2011.
Abstract
Relapsed acute lymphoblastic leukaemia (ALL) is a leading cause of death due to disease in young people, but the biological determinants of treatment failure remain poorly understood. Recent genome-wide profiling of structural DNA alterations in ALL have identified multiple submicroscopic somatic mutations targeting key cellular pathways, and have demonstrated substantial evolution in genetic alterations from diagnosis to relapse. However, DNA sequence mutations in ALL have not been analysed in detail. To identify novel mutations in relapsed ALL, we resequenced 300 genes in matched diagnosis and relapse samples from 23 patients with ALL. This identified 52 somatic non-synonymous mutations in 32 genes, many of which were novel, including the transcriptional coactivators CREBBP and NCOR1, the transcription factors ERG, SPI1, TCF4 and TCF7L2, components of the Ras signalling pathway, histone genes, genes involved in histone modification (CREBBP and CTCF), and genes previously shown to be targets of recurring DNA copy number alteration in ALL. Analysis of an extended cohort of 71 diagnosis-relapse cases and 270 acute leukaemia cases that did not relapse found that 18.3% of relapse cases had sequence or deletion mutations of CREBBP, which encodes the transcriptional coactivator and histone acetyltransferase CREB-binding protein (CREBBP, also known as CBP). The mutations were either present at diagnosis or acquired at relapse, and resulted in truncated alleles or deleterious substitutions in conserved residues of the histone acetyltransferase domain. Functionally, the mutations impaired histone acetylation and transcriptional regulation of CREBBP targets, including glucocorticoid responsive genes. Several mutations acquired at relapse were detected in subclones at diagnosis, suggesting that the mutations may confer resistance to therapy. These results extend the landscape of genetic alterations in leukaemia, and identify mutations targeting transcriptional and epigenetic regulation as a mechanism of resistance in ALL.
Figures
FIGURE 1
CREBBP sequence mutations in relapsed ALL. a, Most variants are missense mutations in CREBBP domains involved in histone acetylation or transcription factor recruitment, or result in protein truncation. NRID, nuclear-receptor-interaction domain; TAZ1/2, transcriptional-adaptor zinc-finger 1/2; KIX, KID-binding domain; Bromo, bromodomain; HAT, histone acetyltransferase domain; ZZ, zinc-binding domain near the dystrophin WW domain; NCBD, nuclear-receptor coactivator-binding domain. b, The locations of CREBBP HAT mutations are shown using the crystal structure of the EP300 HAT domain complexed with its bisubstrate inhibitor, Lys-CoA (blue). CREBBP R1446 (equivalent to EP300 R1410) contacts phosphates of the CoA moiety of the inhibitor (salt bridges are shown as dashed lines), and the R1446H mutation is predicted to disrupt substrate binding. Q1500P (EP300 Q1464) is predicted to disrupt the alpha 4 helix, which stabilizes the substrate binding loop L1 between the beta 5 strand and the alpha 4 helix. C1408H (EP300 C1372) is predicted to disrupt the hydrophobic core that involves both the N and C termini of the HAT domain. R1563 cannot be shown as this residue lies in a proteolytically sensitive autoacetylation loop that was deleted in order to generate the crystal structure. c, Duplication of the R1446H mutation at relapse (R). This mutation was heterozygous at diagnosis (D) and absent in the matched normal sample (N). There is copy neutral loss of heterozygosity of 16p at relapse but not at diagnosis. d, CREBBP mutations are present in subclones at diagnosis, and emerge in the predominant clone at relapse. The S1761* mutation is heterozygous in the relapse sample, absent in the matched normal sample, and appears as a minor peak in the diagnosis sample. Presence of this mutation in a subpopulation of cells at diagnosis was confirmed by PCR, cloning and bidirectional sequencing of multiple colonies of the diagnosis sample (data not shown).
FIGURE 2
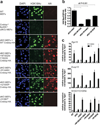
CREBBP mutations impair histone acetylation and multiple gene expression programs. a, Immunofluorescence to detect histone H3 lysine 18 acetylation (H3K18Ac) in nontransduced wild type MEFs, CrebbpΔflox/Δflox;Ep300Δflox/Δflox (dKO) MEFs and dKO MEFs transduced with retrovirus expressing wild type (+Crebbp) and mutant HA-tagged Crebbp. Four independent experiments with separate controls were performed, one of which is shown. S1887* truncates the protein before the C-terminal HA-tag, but is predicted to retain the HAT domain. b, Quantification of H3K18Ac mean signal intensity per nucleus relative to the HA-tagged Crebbp retrovirus mean signal intensity. Mean +/− SEM; 40–61 nuclei quantified per retrovirus. Only nuclei that have an HA-tag (Crebbp-HA) signal greater than 2.5 fold above background were included. Data is expressed as the ratio of the mean H3K18Ac signal intensity for each nucleus to the mean HA signal intensity for the same nuclei. P value shown is from Tukey post test of one way ANOVA. c, Quantitative RT-PCR gene expression data from dKO MEFs transduced with wild type or mutant CBP treated for 90 min with 10 µM forskolin + 100µM IBMX (FI). W1502A/Y1503S is a previously described dominant negative mutation. Gene expression was normalized to expression of Pgk1. N=3–7.
Similar articles
- Inactivating mutations of acetyltransferase genes in B-cell lymphoma.
Pasqualucci L, Dominguez-Sola D, Chiarenza A, Fabbri G, Grunn A, Trifonov V, Kasper LH, Lerach S, Tang H, Ma J, Rossi D, Chadburn A, Murty VV, Mullighan CG, Gaidano G, Rabadan R, Brindle PK, Dalla-Favera R. Pasqualucci L, et al. Nature. 2011 Mar 10;471(7337):189-95. doi: 10.1038/nature09730. Nature. 2011. PMID: 21390126 Free PMC article. - CREBBP knockdown enhances RAS/RAF/MEK/ERK signaling in Ras pathway mutated acute lymphoblastic leukemia but does not modulate chemotherapeutic response.
Dixon ZA, Nicholson L, Zeppetzauer M, Matheson E, Sinclair P, Harrison CJ, Irving JA. Dixon ZA, et al. Haematologica. 2017 Apr;102(4):736-745. doi: 10.3324/haematol.2016.145177. Epub 2016 Dec 15. Haematologica. 2017. PMID: 27979926 Free PMC article. - Mutations in epigenetic regulators including SETD2 are gained during relapse in paediatric acute lymphoblastic leukaemia.
Mar BG, Bullinger LB, McLean KM, Grauman PV, Harris MH, Stevenson K, Neuberg DS, Sinha AU, Sallan SE, Silverman LB, Kung AL, Lo Nigro L, Ebert BL, Armstrong SA. Mar BG, et al. Nat Commun. 2014 Mar 24;5:3469. doi: 10.1038/ncomms4469. Nat Commun. 2014. PMID: 24662245 Free PMC article. - Rubinstein-Taybi Syndrome and Epigenetic Alterations.
Korzus E. Korzus E. Adv Exp Med Biol. 2017;978:39-62. doi: 10.1007/978-3-319-53889-1_3. Adv Exp Med Biol. 2017. PMID: 28523540 Free PMC article. Review. - Dysregulation of the p300/CBP histone acetyltransferases in human cancer.
Xu L, Xuan H, Shi X. Xu L, et al. Epigenomics. 2025 Feb;17(3):193-208. doi: 10.1080/17501911.2024.2447807. Epub 2024 Dec 30. Epigenomics. 2025. PMID: 39929233 Review.
Cited by
- Kinase pathway dependence in primary human leukemias determined by rapid inhibitor screening.
Tyner JW, Yang WF, Bankhead A 3rd, Fan G, Fletcher LB, Bryant J, Glover JM, Chang BH, Spurgeon SE, Fleming WH, Kovacsovics T, Gotlib JR, Oh ST, Deininger MW, Zwaan CM, Den Boer ML, van den Heuvel-Eibrink MM, O'Hare T, Druker BJ, Loriaux MM. Tyner JW, et al. Cancer Res. 2013 Jan 1;73(1):285-96. doi: 10.1158/0008-5472.CAN-12-1906. Epub 2012 Oct 18. Cancer Res. 2013. PMID: 23087056 Free PMC article. - Attenuation of AML1-ETO cellular dysregulation correlates with increased leukemogenic potential.
DeKelver RC, Yan M, Ahn EY, Shia WJ, Speck NA, Zhang DE. DeKelver RC, et al. Blood. 2013 May 2;121(18):3714-7. doi: 10.1182/blood-2012-11-465641. Epub 2013 Feb 20. Blood. 2013. PMID: 23426948 Free PMC article. - Role of epigenetic in cancer biology, in hematologic malignancies and in anticancer therapy.
Nwabo Kamdje AH, Dongmo Fogang HP, Mimche PN. Nwabo Kamdje AH, et al. Front Mol Med. 2024 Sep 6;4:1426454. doi: 10.3389/fmmed.2024.1426454. eCollection 2024. Front Mol Med. 2024. PMID: 39308891 Free PMC article. Review. - Interplay between the cancer genome and epigenome.
Shen H, Laird PW. Shen H, et al. Cell. 2013 Mar 28;153(1):38-55. doi: 10.1016/j.cell.2013.03.008. Cell. 2013. PMID: 23540689 Free PMC article. Review. - Clonal evolution of the 3D chromatin landscape in patients with relapsed pediatric B-cell acute lymphoblastic leukemia.
Narang S, Ghebrechristos Y, Evensen NA, Murrell N, Jasinski S, Ostrow TH, Teachey DT, Raetz EA, Lionnet T, Witkowski M, Aifantis I, Tsirigos A, Carroll WL. Narang S, et al. Nat Commun. 2024 Aug 28;15(1):7425. doi: 10.1038/s41467-024-51492-6. Nat Commun. 2024. PMID: 39198446 Free PMC article.
References
- Mullighan CG, et al. Genome-wide analysis of genetic alterations in acute lymphoblastic leukaemia. Nature. 2007;446:758–764. - PubMed
- Kuiper RP, et al. High-resolution genomic profiling of childhood ALL reveals novel recurrent genetic lesions affecting pathways involved in lymphocyte differentiation and cell cycle progression. Leukemia. 2007;21:1258–1266. - PubMed
- Goodman RH, Smolik S. CBP/p300 in cell growth, transformation, and development. Genes Dev. 2000;14:1553–1577. - PubMed
- Pui CH, Robison LL, Look AT. Acute lymphoblastic leukaemia. Lancet. 2008;371:1030–1043. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials