Antibiotic resistance genes in the bacteriophage DNA fraction of environmental samples - PubMed (original) (raw)
Antibiotic resistance genes in the bacteriophage DNA fraction of environmental samples
Marta Colomer-Lluch et al. PLoS One. 2011.
Abstract
Antibiotic resistance is an increasing global problem resulting from the pressure of antibiotic usage, greater mobility of the population, and industrialization. Many antibiotic resistance genes are believed to have originated in microorganisms in the environment, and to have been transferred to other bacteria through mobile genetic elements. Among others, β-lactam antibiotics show clinical efficacy and low toxicity, and they are thus widely used as antimicrobials. Resistance to β-lactam antibiotics is conferred by β-lactamase genes and penicillin-binding proteins, which are chromosomal- or plasmid-encoded, although there is little information available on the contribution of other mobile genetic elements, such as phages. This study is focused on three genes that confer resistance to β-lactam antibiotics, namely two β-lactamase genes (blaTEM and blaCTX-M9) and one encoding a penicillin-binding protein (mecA) in bacteriophage DNA isolated from environmental water samples. The three genes were quantified in the DNA isolated from bacteriophages collected from 30 urban sewage and river water samples, using quantitative PCR amplification. All three genes were detected in the DNA of phages from all the samples tested, in some cases reaching 104 gene copies (GC) of blaTEM or 102 GC of blaCTX-M and mecA. These values are consistent with the amount of fecal pollution in the sample, except for mecA, which showed a higher number of copies in river water samples than in urban sewage. The bla genes from phage DNA were transferred by electroporation to sensitive host bacteria, which became resistant to ampicillin. blaTEM and blaCTX were detected in the DNA of the resistant clones after transfection. This study indicates that phages are reservoirs of resistance genes in the environment.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
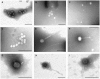
Figure 1. Electron micrographs of bacteriophages present in sewage and river water.
A–B. Group of phages with Myoviridae and Siphoviridae morphology from sewage. C. Myoviridae phages from river water. D: group of Siphoviridae phages from sewage. E–F. Myoviridae phages from sewage. G: Podoviridae phage from sewage. H–I. Siphoviridae phages from sewage and river water respectively. Bar 200 nm.
Figure 2. Number of copies of bla TEM genes (GC/ml) in urban sewage and river water samples in phage and bacterial DNA.
On the left side of the figure, bar chart of the gene copies detected for each sample, dark grey for phage DNA and light grey for bacterial DNA. On the right side of the figure, the box plot chart shows the averaged values obtained from all samples from the same origin. Within the box plot chart, the cross-pieces of each box plot represent (from top to bottom) maximum, upper-quartile, median (black bar), lower-quartile, and minimum values. Black diamond shows the mean value. The grey boxes in the box plot chart include samples showing values within the 75th percentile and white boxes samples showing values within the within the 25th percentile.
Figure 3. Primers and TaqMan assay probe resulting from the alignment of bla CTX-M genes from Cluster 1.
Conserved nucleotides are marked in bold, non-conserved nucleotides in white. Sequence reverse and complementary is shown for lower primer. Right column indicate the GenBank accession number of each gene.
Figure 4. Number of copies of bla CTX-M genes (GC/ml) in urban sewage and river water samples in phage and bacterial DNA and box plot of averaged values.
Figure 5. Number of copies of _mec_A (GC/ml) in urban sewage and river water samples in phage and bacterial DNA and box plot of averaged values.
Similar articles
- Sludge as a potential important source of antibiotic resistance genes in both the bacterial and bacteriophage fractions.
Calero-Cáceres W, Melgarejo A, Colomer-Lluch M, Stoll C, Lucena F, Jofre J, Muniesa M. Calero-Cáceres W, et al. Environ Sci Technol. 2014 Jul 1;48(13):7602-11. doi: 10.1021/es501851s. Epub 2014 Jun 10. Environ Sci Technol. 2014. PMID: 24873655 - Contribution of bacteriophage and plasmid DNA to the mobilization of antibiotic resistance genes in a river receiving treated wastewater discharges.
Lekunberri I, Villagrasa M, Balcázar JL, Borrego CM. Lekunberri I, et al. Sci Total Environ. 2017 Dec 1;601-602:206-209. doi: 10.1016/j.scitotenv.2017.05.174. Epub 2017 May 25. Sci Total Environ. 2017. PMID: 28551539 - Antibiotic resistance genes in the bacteriophage DNA fraction of human fecal samples.
Quirós P, Colomer-Lluch M, Martínez-Castillo A, Miró E, Argente M, Jofre J, Navarro F, Muniesa M. Quirós P, et al. Antimicrob Agents Chemother. 2014;58(1):606-9. doi: 10.1128/AAC.01684-13. Epub 2013 Oct 28. Antimicrob Agents Chemother. 2014. PMID: 24165177 Free PMC article. - Plasmids in the environment.
Linton AH. Linton AH. Schriftenr Ver Wasser Boden Lufthyg. 1988;78:197-224. Schriftenr Ver Wasser Boden Lufthyg. 1988. PMID: 3074480 Review. - Bacteriophages as Environmental Reservoirs of Antibiotic Resistance.
Calero-Cáceres W, Ye M, Balcázar JL. Calero-Cáceres W, et al. Trends Microbiol. 2019 Jul;27(7):570-577. doi: 10.1016/j.tim.2019.02.008. Epub 2019 Mar 21. Trends Microbiol. 2019. PMID: 30905524 Review.
Cited by
- Human health implications of clinically relevant bacteria in wastewater habitats.
Varela AR, Manaia CM. Varela AR, et al. Environ Sci Pollut Res Int. 2013 Jun;20(6):3550-69. doi: 10.1007/s11356-013-1594-0. Epub 2013 Mar 19. Environ Sci Pollut Res Int. 2013. PMID: 23508533 Review. - How Broad Is Enough: The Host Range of Bacteriophages and Its Impact on the Agri-Food Sector.
Fong K, Wong CWY, Wang S, Delaquis P. Fong K, et al. Phage (New Rochelle). 2021 Jun 1;2(2):83-91. doi: 10.1089/phage.2020.0036. Epub 2021 Jun 16. Phage (New Rochelle). 2021. PMID: 36148040 Free PMC article. Review. - Extensive antimicrobial resistance mobilization via multicopy plasmid encapsidation mediated by temperate phages.
Rodríguez-Rubio L, Serna C, Ares-Arroyo M, Matamoros BR, Delgado-Blas JF, Montero N, Bernabe-Balas C, Wedel EF, Mendez IS, Muniesa M, Gonzalez-Zorn B. Rodríguez-Rubio L, et al. J Antimicrob Chemother. 2020 Nov 1;75(11):3173-3180. doi: 10.1093/jac/dkaa311. J Antimicrob Chemother. 2020. PMID: 32719862 Free PMC article. - Biogeographical variation in antimicrobial resistance in rivers is influenced by agriculture and is spread through bacteriophages.
Andersson T, Adell AD, Moreno-Switt AI, Spégel P, Turner C, Overballe-Petersen S, Fuursted K, Lood R. Andersson T, et al. Environ Microbiol. 2022 Oct;24(10):4869-4884. doi: 10.1111/1462-2920.16122. Epub 2022 Jul 7. Environ Microbiol. 2022. PMID: 35799549 Free PMC article. - Bacteriophages and genetic mobilization in sewage and faecally polluted environments.
Muniesa M, Imamovic L, Jofre J. Muniesa M, et al. Microb Biotechnol. 2011 Nov;4(6):725-34. doi: 10.1111/j.1751-7915.2011.00264.x. Epub 2011 Apr 27. Microb Biotechnol. 2011. PMID: 21535427 Free PMC article. Review.
References
- World Health Organization. Geneva, Switzerland.: 1996. The world health report.
- American Academy of Microbiology. ASM. Washington; 2009. Antibiotic Resistance: An Ecological Perspective on an Old Problem.
- Hawkey PM, Jones AM. The changing epidemiology of resistance. J Antimicrob Chemother. 2009;64S1:i3–10. - PubMed
- Canton R. Antibiotic resistance genes from the environment: a perspective through newly identified antibiotic resistance mechanisms in the clinical setting. Clin Microbiol Infect. 2009;1:20–25. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous