Population-genetic properties of differentiated human copy-number polymorphisms - PubMed (original) (raw)
Population-genetic properties of differentiated human copy-number polymorphisms
Catarina D Campbell et al. Am J Hum Genet. 2011.
Abstract
Copy-number variants (CNVs) can reach appreciable frequencies in the human population, and recent discoveries have shown that several of these copy-number polymorphisms (CNPs) are associated with human diseases, including lupus, psoriasis, Crohn disease, and obesity. Despite new advances, significant biases remain in terms of CNP discovery and genotyping. We developed a method based on single-channel intensity data and benchmarked against copy numbers determined from sequencing read depth to successfully obtain CNP genotypes for 1495 CNPs from 487 human DNA samples of diverse ethnic backgrounds. This microarray contained CNPs in segmental duplication-rich regions and insertions of sequences not represented in the reference genome assembly or on standard SNP microarray platforms. We observe that CNPs in segmental duplications are more likely to be population differentiated than CNPs in unique regions (p = 0.015) and that biallelic CNPs show greater stratification when compared to frequency-matched SNPs (p = 0.0026). Although biallelic CNPs show a strong correlation of copy number with flanking SNP genotypes, the majority of multicopy CNPs do not (40% with r > 0.8). We selected a subset of CNPs for further characterization in 1876 additional samples from 62 populations; this revealed striking population-differentiated structural variants in genes of clinical significance such as OCLN, a tight junction protein involved in hepatitis C viral entry. Our microarray design allows these variants to be rapidly tested for disease association and our results suggest that CNPs (especially those that cannot be imputed from SNP genotypes) might have contributed disproportionately to human diversity and selection.
Copyright © 2011 The American Society of Human Genetics. Published by Elsevier Inc. All rights reserved.
Figures
Figure 1
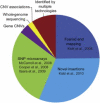
Targeted Copy-Number Polymorphisms A pie chart of the sources for the 4041 targeted CNPs.
Figure 2
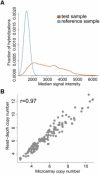
Using Array CGH to Estimate Copy Numbers for Loci without Discrete Copy-Number Classes (A) Distributions of single-channel intensity values for the test sample (orange) and the reference sample (blue). The reference sample shows high reproducibility across all microarrays. Because this is a CNP, the test samples show much more variability in single-channel intensity. (B) Copy number determined from single-channel intensity data are highly correlated to sequencing read depth copy-number estimates for a CNP overlapping NPEPPS on chromosome 17. The fit of this line may be used for subsequent determinations of copy number.
Figure 3
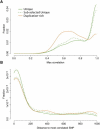
CNPs in SDs Show Less LD to SNPs than CNPs in Unique Regions (A) The distribution of correlation coefficients between copy number and SNP genotype are shown for CNPs in SDs (orange) and CNPs in unique regions (green). The dashed line represents the average maximum correlation across 100 samplings of the CNPs in unique regions to match the distances to the most correlated SNP for CNPs in duplication-rich regions. All SNPs within 1 Mb of the CNP were tested in five populations (European American [CEU], Han Chinese from Beijing [CHB], Japanese [JPT], Maasai [MKK], and Yoruba [YRI]) and the highest correlation coefficient in all populations was included. (B) Distributions of the distance from the CNP to the most correlated SNP. The distance is slightly larger for CNPs in SDs (p = 0.3), but this does not explain the large difference in LD.
Figure 4
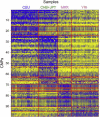
Population Differentiation of CNPs with High VST Values The top 100 CNPs based on maximum VST between all pairwise comparisons of populations are shown for the initial analysis in 487 individuals from five populations: European American (CEU), Han Chinese from Beijing (CHB), Japanese (JPT), Maasai (MKK), and Yoruba (YRI). Blue color in the heatmap represents reduced copy when compared to the reference sample (a CEU female) and yellow represents increased copy number. Loci are clustered based on the pattern of hybridization values across populations.
Figure 5
Comparisons of Population Differentiation between Different Classes of Variants Histograms of VST or FST values are plotted. (A) Informative CNPs were stratified based on their duplication content; CNPs with at least 50% overlap with SDs or regions of excess read depth in the Celera genome were defined as duplication rich. CNPs with zero bases of SD or excess read depth were defined as unique. Distributions of maximum VST value for each CNP are plotted for both classes of variants. These distributions are significantly different from one another (Kolmogorov-Smirnov two-tailed test, p = 0.015). (B) Comparison of FST statistics for biallelic autosomal CNPs compared to frequency-matched, autosomal SNPs (Kolmogorov-Smirnov two-tailed test, p = 0.0026).
Figure 6
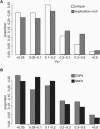
Examples of Population-Differentiated Loci Histograms of log2 ratios are plotted for the unrelated individuals in each population. (A) Diagram of the bitter taste receptor cluster on chromosome 12 and distribution of log2 ratios for a CNP containing TAS2R46. The maximum VST is 0.63 between YRI and JPT. (B) Diagram of the CNP containing the last five exons of OCLN and the distribution of log2 ratios for a CNP in OCLN. The maximum VST for this locus is 0.51 between YRI and CHB.
Figure 7
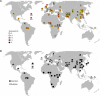
Worldwide Distributions of Selected CNPs We designed PCR or qPCR assays to genotype selected CNPs in HGDP individuals from 52 populations. Included in the figure are the copy-number distributions for the 12 populations tested with microarray. These pie charts are labeled with population codes. (A) We obtained copy-number estimates from qPCR for 687 individuals for the CNP overlapping OCLN. The distributions of estimated copy number for each population with data in at least five individuals are overlaid on a map of the world. (B) We obtained allele frequencies for an insertion of novel sequence located near ATP6V1G3 for 952 HGDP individuals. The allele frequencies of the insertion (black) and the deletion allele (white) are shown for each population.
Similar articles
- Founder population-specific HapMap panel increases power in GWA studies through improved imputation accuracy and CNV tagging.
Surakka I, Kristiansson K, Anttila V, Inouye M, Barnes C, Moutsianas L, Salomaa V, Daly M, Palotie A, Peltonen L, Ripatti S. Surakka I, et al. Genome Res. 2010 Oct;20(10):1344-51. doi: 10.1101/gr.106534.110. Epub 2010 Sep 1. Genome Res. 2010. PMID: 20810666 Free PMC article. - Linkage disequilibrium and heritability of copy-number polymorphisms within duplicated regions of the human genome.
Locke DP, Sharp AJ, McCarroll SA, McGrath SD, Newman TL, Cheng Z, Schwartz S, Albertson DG, Pinkel D, Altshuler DM, Eichler EE. Locke DP, et al. Am J Hum Genet. 2006 Aug;79(2):275-90. doi: 10.1086/505653. Epub 2006 Jun 15. Am J Hum Genet. 2006. PMID: 16826518 Free PMC article. - Molecular methods for genotyping complex copy number polymorphisms.
Cantsilieris S, Baird PN, White SJ. Cantsilieris S, et al. Genomics. 2013 Feb;101(2):86-93. doi: 10.1016/j.ygeno.2012.10.004. Epub 2012 Oct 30. Genomics. 2013. PMID: 23123317 Review. - Copy-number variation and association studies of human disease.
McCarroll SA, Altshuler DM. McCarroll SA, et al. Nat Genet. 2007 Jul;39(7 Suppl):S37-42. doi: 10.1038/ng2080. Nat Genet. 2007. PMID: 17597780 Review.
Cited by
- Structurally divergent and recurrently mutated regions of primate genomes.
Mao Y, Harvey WT, Porubsky D, Munson KM, Hoekzema K, Lewis AP, Audano PA, Rozanski A, Yang X, Zhang S, Yoo D, Gordon DS, Fair T, Wei X, Logsdon GA, Haukness M, Dishuck PC, Jeong H, Del Rosario R, Bauer VL, Fattor WT, Wilkerson GK, Mao Y, Shi Y, Sun Q, Lu Q, Paten B, Bakken TE, Pollen AA, Feng G, Sawyer SL, Warren WC, Carbone L, Eichler EE. Mao Y, et al. Cell. 2024 Mar 14;187(6):1547-1562.e13. doi: 10.1016/j.cell.2024.01.052. Epub 2024 Feb 29. Cell. 2024. PMID: 38428424 - Genomic structural variation: A complex but important driver of human evolution.
Soto DC, Uribe-Salazar JM, Shew CJ, Sekar A, McGinty SP, Dennis MY. Soto DC, et al. Am J Biol Anthropol. 2023 Aug;181 Suppl 76(Suppl 76):118-144. doi: 10.1002/ajpa.24713. Epub 2023 Feb 16. Am J Biol Anthropol. 2023. PMID: 36794631 Free PMC article. Review. - Frequent copy number variants in a cohort of Mexican-Mestizo individuals.
Sánchez S, Juárez U, Domínguez J, Molina B, Barrientos R, Martínez-Hernández A, Carnevale A, Grether-González P, Mayen DG, Villarroel C, Lieberman E, Yokoyama E, Del Castillo V, Torres L, Frias S. Sánchez S, et al. Mol Cytogenet. 2023 Jan 12;16(1):2. doi: 10.1186/s13039-022-00631-z. Mol Cytogenet. 2023. PMID: 36631885 Free PMC article. - The individual and global impact of copy-number variants on complex human traits.
Auwerx C, Lepamets M, Sadler MC, Patxot M, Stojanov M, Baud D, Mägi R; Estonian Biobank Research Team; Porcu E, Reymond A, Kutalik Z. Auwerx C, et al. Am J Hum Genet. 2022 Apr 7;109(4):647-668. doi: 10.1016/j.ajhg.2022.02.010. Epub 2022 Mar 2. Am J Hum Genet. 2022. PMID: 35240056 Free PMC article. - Quantitative assessment reveals the dominance of duplicated sequences in germline-derived extrachromosomal circular DNA.
Mouakkad-Montoya L, Murata MM, Sulovari A, Suzuki R, Osia B, Malkova A, Katsumata M, Giuliano AE, Eichler EE, Tanaka H. Mouakkad-Montoya L, et al. Proc Natl Acad Sci U S A. 2021 Nov 23;118(47):e2102842118. doi: 10.1073/pnas.2102842118. Proc Natl Acad Sci U S A. 2021. PMID: 34789574 Free PMC article.
References
- Sebat J., Lakshmi B., Troge J., Alexander J., Young J., Lundin P., Månér S., Massa H., Walker M., Chi M. Large-scale copy number polymorphism in the human genome. Science. 2004;305:525–528. - PubMed
- Iafrate A.J., Feuk L., Rivera M.N., Listewnik M.L., Donahoe P.K., Qi Y., Scherer S.W., Lee C. Detection of large-scale variation in the human genome. Nat. Genet. 2004;36:949–951. - PubMed
- Hinds D.A., Kloek A.P., Jen M., Chen X., Frazer K.A. Common deletions and SNPs are in linkage disequilibrium in the human genome. Nat. Genet. 2006;38:82–85. - PubMed
- Conrad D.F., Andrews T.D., Carter N.P., Hurles M.E., Pritchard J.K. A high-resolution survey of deletion polymorphism in the human genome. Nat. Genet. 2006;38:75–81. - PubMed
- McCarroll S.A., Hadnott T.N., Perry G.H., Sabeti P.C., Zody M.C., Barrett J.C., Dallaire S., Gabriel S.B., Lee C., Daly M.J., Altshuler D.M., International HapMap Consortium Common deletion polymorphisms in the human genome. Nat. Genet. 2006;38:86–92. - PubMed
Publication types
MeSH terms
Grants and funding
- F32HG006070/HG/NHGRI NIH HHS/United States
- T32HG00035/HG/NHGRI NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- T32 HG000035/HG/NHGRI NIH HHS/United States
- HG004120/HG/NHGRI NIH HHS/United States
- F32 HG006070/HG/NHGRI NIH HHS/United States
- P01 HG004120/HG/NHGRI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous