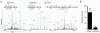Phosphoproteomic analysis reveals an intrinsic pathway for the regulation of histone deacetylase 7 that controls the function of cytotoxic T lymphocytes - PubMed (original) (raw)
Phosphoproteomic analysis reveals an intrinsic pathway for the regulation of histone deacetylase 7 that controls the function of cytotoxic T lymphocytes
Maria N Navarro et al. Nat Immunol. 2011 Apr.
Abstract
Here we report an unbiased analysis of the cytotoxic T lymphocyte (CTL) serine-threonine phosphoproteome by high-resolution mass spectrometry. We identified approximately 2,000 phosphorylations in CTLs, of which approximately 450 were controlled by T cell antigen receptor (TCR) signaling. A significantly overrepresented group of molecules identified included transcription activators, corepressors and chromatin regulators. A focus on chromatin regulators showed that CTLs had high expression of the histone deacetylase HDAC7 but continually phosphorylated and exported this transcriptional repressor from the nucleus. Dephosphorylation of HDAC7 resulted in its accumulation in the nucleus and suppressed expression of genes encoding key cytokines, cytokine receptors and adhesion molecules that determine CTL function. Screening of the CTL phosphoproteome has thus identified intrinsic pathways of serine-threonine phosphorylation that target chromatin regulators and determine the CTL functional program.
Figures
Figure 1
Analysis of the basal and TCR regulated phosphoproteome in CTLs. P14 LCMV CTLs differentially labeled in SILAC media were either left unstimulated or triggered via their TCR with cognate peptide for 1 h. Cells were lysed and phosphopeptide purification using HILIC/IMAC enrichment was performed as described in Supplementary Methods. The resulting peptides were analyzed and identified using a LTQ-Orbitrap XL via MaxQuant v.1.0.13.13. (a) Graphic shows the ratio/ intensity plot of 2,078 phosphopeptides identified in T cell lysates. TCR downregulated and upregulated phosphorylations are indicated as black dots. Selected proteins that have been previously shown to change phosphorylation after TCR stimulation are indicated. (b) All 955 identified phosphoproteins were subjected Ingenuity Pathway Analysis. The result of the molecular and cellular function analysis is shown.
Figure 2
Ingenuity Pathway Analysis of consistent phosphorylations in CTLs. SILAC and HILIC/IMAC purification protocols were independently performed four times, including amino acid labeling switch. The isotope combination (unstimulated/stimulated) was as follows: experiment 1, R10K8/R0K0 (Fig. 1), experiment 2, R0K0/R6K6, experiment 3 and 4, R0K0/R6K6 (not shown). CTLs were stimulated for 1 h with cognate peptide in experiments 1, 2 and 3 and for 10 min in experiment 4. Reproducible phosphorylations in three out of four experiments were considered for analysis. (a) Ingenuity Pathway Analysis of 742 phosphorylations on 473 different proteins found in at least three of the four conducted SILAC experiments. (b) Ingenuity Pathway Analysis of TCR-regulated 94 phosphorylations on 78 different proteins consistently found in our screening using 1.5 fold as threshold for regulation. (c) Representation of the frequency of kinases predicted to be active in CTL using MaxQuant software analysis. MaxQuant uses a sequence window ± 6 amino acids around the identified phosphorylation site to determine the kinase that could phosphorylate this motif.
Figure 3
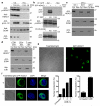
Phosphorylated chromatin regulators in CTLs: class II histone deacetylase 7 (HDAC7). (a) De novo synthesis of the acquired pseudo MS3 spectra in one of the four experiments performed for the three peptides KTVpSEPNLK, KEpSAPPSLR and pTRSEPLPPSATASPLLAPLQPR. * in y and b ion series indicates loss of phosphate. (b) Spectral counting of the two detected class IIa HDACs in CTLs, HDAC7 and HDAC4 separately calculated for phosphopeptide enrichment and 14-3-3 affinity purification screens. Graph represents the averaged spectral counts of the four SILAC experiments ± SEM.
Figure 4
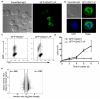
Subcellular distribution of HDAC7 in CTLs. (a) Immunoblot analysis of CTLs cytosolic and nuclear extracts treated with peptide (TCR), PDBu (PD) or untreated (−) for 30 min. Anti-IκBα and anti-SCM1 were included as controls for fraction purity and anti-p-Erk as activation control. Representative of 3 experiments. (b) Immunoblot analysis of CTLs retrovirally transduced with GFP-HDAC7. (Top) cytosolic and nuclear extracts from GFP-HDAC7 transduced (+) or non-transduced (−) were immunoprecipitated and immunoblotted with anti-GFP. (Middle, bottom), GFP-HDAC7 transduced CTLs were stimulated with PDBu (PD), peptide (TCR) or unstimulated (−) for 2h. In middle panel, cytosolic extracts were immunoprecipitated with anti-GFP and immunoblotted with anti-p-HDAC7. In bottom panel, cytosolic extracts were affinity purified with 14-3-3-sepharose and immunoblotted with anti-GFP. Data are representative of two experiments. (c) Immunoblot analysis of HDAC7 expression in cytosolic and nuclear extracts of CTLs obtained from OT1 and two sets of polyclonal, non-TCR-Tg mice (WT). Representative of 2 experiments. (d) Immunoblot analysis of HDAC7 expression in cytosolic and nuclear extracts of different populations of naïve T cells: CD8+ cells from TCR-Tg OT1 mice, polyclonal CD4+ and total T cells from non-TCR-Tg (WT). Representative of 2 experiments. (e) P14-LCMV CTLs retrovirally transduced with a vector encoding GFP-HDAC7 chimeric protein were analyzed by confocal microscopy. Images are representative of ten independent experiments. (f) GFP-HDAC7 expressing P14-LCMV CTLs were left untreated (U) or treated with leptomycin B (L) and stained with DAPI. Images are representative of 3h of LMB treatment. First graph represents the percentage of cells with nuclear GFP-HDAC7 at different time points of LMB treatment. Second graph represents average ± SEM after 3h of LMB (_n_=7). At least 100 cells were counted in each experiment and time point.
Figure 5
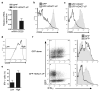
HDAC7 nuclear exclusion is required for normal CTL function. P14-LCMV CTLs were retrovirally transduced with GFP-HDAC7-ΔP or GFP-HDAC7. In (a) and (b) subcellular distribution of GFP-HDAC7-ΔP was analyzed by microscopy. In (b), DAPI staining was included. Images are representative of 4 experiments. (c) GFP-HDAC7 and GFP-HDAC7-ΔP transduced CTLs were analyzed by flow cytometry. Plots representative of 10 experiments. (d) Cell numbers and GFP expression were assessed daily and represented as cell number over time. Graph represents average number ± SEM of 3 experiments. (e) GFP-HDAC7-ΔP transduced CTLs were sorted based on GFP expression, and microarray analysis was performed using Affymetrix GeneChip mouse genome 430_2.0 array comparing expression profile of GFP-HDAC7-ΔP (GFP+) and control (GFP−) cells. Graph shows the distribution of the intensity ratio (log2 fold change, GFP-HDAC7-ΔP relative to control) plotted by the average of the normalized intensity values for 23,653 probes identified as present in at least one sample. Probes with 2 fold change or higher are represented by black dots (1,457 probes, 993 annotated genes). Grey dots represent probes with no significant change or fold change below 2 fold (22,196 probes, 11,148 annotated genes). Using a 2-fold cut, 265 genes show decreased expression in GFP-HDAC7-ΔP cells, and 728 genes shown increased expression. Data are accessible through GEO Series accession number GSE27092 (
http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE27092
).
Figure 6
HDAC7 nuclear exclusion is required for expression of the high affinity IL2 receptor. (a) Relative expression of CD25 mRNA in sorted GFP negative (GFP−) and GFP-HDAC7-ΔP. Data shown are an average of three different experiments ± SEM (*AU= arbitrary units). (b) GFP-HDAC7-ΔP transduced P14-LCMV CTLs were stained for CD25 and analyzed by flow cytometry. GFP positive and negative cells were electronically gated to compare CD25 expression in both populations. (c) GFP-HDAC7 and GFP-HDAC7-ΔP transduced P14-LCMV-CTLs were stained for CD25 and analyzed by flow cytometry. Histograms in b and c are representative of three independently performed experiments. (d,e) P14-LCMV CTLs were stimulated with cognate peptide for 4h before assessing surface CD25 and intracellular IFN-γ expression by flow cytometry. (e) Expression of CD25 was electronically gated as in (d) to compare IFN-γ expression. The percentage of IFN-γ positive cells in both populations is represented as an averaged value of four experiments ± SEM. (f) IFN-γ production was assessed by intracellular staining in P14-LCMV CTLs expressing GFP-HDAC7-ΔP or GFP alone after stimulation with peptide for 4h. Data are representative of three experiments.
Comment in
- To kill, you have to duck an HDAC.
Altman A, Kong KF. Altman A, et al. Nat Immunol. 2011 Apr;12(4):279-81. doi: 10.1038/ni0411-279. Nat Immunol. 2011. PMID: 21423220 No abstract available.
Similar articles
- To kill, you have to duck an HDAC.
Altman A, Kong KF. Altman A, et al. Nat Immunol. 2011 Apr;12(4):279-81. doi: 10.1038/ni0411-279. Nat Immunol. 2011. PMID: 21423220 No abstract available. - Histone deacetylase 7 regulates cell survival and TCR signaling in CD4/CD8 double-positive thymocytes.
Kasler HG, Young BD, Mottet D, Lim HW, Collins AM, Olson EN, Verdin E. Kasler HG, et al. J Immunol. 2011 Apr 15;186(8):4782-93. doi: 10.4049/jimmunol.1001179. Epub 2011 Mar 11. J Immunol. 2011. PMID: 21398603 - Quantitative phosphoproteomics of cytotoxic T cells to reveal protein kinase d 2 regulated networks.
Navarro MN, Goebel J, Hukelmann JL, Cantrell DA. Navarro MN, et al. Mol Cell Proteomics. 2014 Dec;13(12):3544-57. doi: 10.1074/mcp.M113.037242. Epub 2014 Sep 29. Mol Cell Proteomics. 2014. PMID: 25266776 Free PMC article. - Phosphorylation of histone deacetylase 7 by protein kinase D mediates T cell receptor-induced Nur77 expression and apoptosis.
Dequiedt F, Van Lint J, Lecomte E, Van Duppen V, Seufferlein T, Vandenheede JR, Wattiez R, Kettmann R. Dequiedt F, et al. J Exp Med. 2005 Mar 7;201(5):793-804. doi: 10.1084/jem.20042034. Epub 2005 Feb 28. J Exp Med. 2005. PMID: 15738054 Free PMC article. - Class II histone deacetylases: structure, function, and regulation.
Bertos NR, Wang AH, Yang XJ. Bertos NR, et al. Biochem Cell Biol. 2001;79(3):243-52. Biochem Cell Biol. 2001. PMID: 11467738 Review.
Cited by
- Combining HDAC inhibitors with oncolytic virotherapy for cancer therapy.
Nakashima H, Nguyen T, Chiocca EA. Nakashima H, et al. Oncolytic Virother. 2015 Nov 20;4:183-91. doi: 10.2147/OV.S66081. eCollection 2015. Oncolytic Virother. 2015. PMID: 27512681 Free PMC article. Review. - Regulation of histone deacetylase activities and functions by phosphorylation and its physiological relevance.
Bahl S, Seto E. Bahl S, et al. Cell Mol Life Sci. 2021 Jan;78(2):427-445. doi: 10.1007/s00018-020-03599-4. Epub 2020 Jul 18. Cell Mol Life Sci. 2021. PMID: 32683534 Free PMC article. Review. - RasGRP Ras guanine nucleotide exchange factors in cancer.
Ksionda O, Limnander A, Roose JP. Ksionda O, et al. Front Biol (Beijing). 2013 Oct 1;8(5):508-532. doi: 10.1007/s11515-013-1276-9. Front Biol (Beijing). 2013. PMID: 24744772 Free PMC article. - Protein kinase D: coupling extracellular stimuli to the regulation of cell physiology.
Fu Y, Rubin CS. Fu Y, et al. EMBO Rep. 2011 Jul 8;12(8):785-96. doi: 10.1038/embor.2011.139. EMBO Rep. 2011. PMID: 21738220 Free PMC article. Review. - A protein phosphatase network controls the temporal and spatial dynamics of differentiation commitment in human epidermis.
Mishra A, Oulès B, Pisco AO, Ly T, Liakath-Ali K, Walko G, Viswanathan P, Tihy M, Nijjher J, Dunn SJ, Lamond AI, Watt FM. Mishra A, et al. Elife. 2017 Oct 18;6:e27356. doi: 10.7554/eLife.27356. Elife. 2017. PMID: 29043977 Free PMC article.
References
- Salmond RJ, Emery J, Okkenhaug K, Zamoyska R. MAPK, phosphatidylinositol 3-kinase, and mammalian target of rapamycin pathways converge at the level of ribosomal protein S6 phosphorylation to control metabolic signaling in CD8 T cells. J Immunol. 2009;183:7388–7397. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases