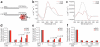Rudimentary G-quadruplex-based telomere capping in Saccharomyces cerevisiae - PubMed (original) (raw)
doi: 10.1038/nsmb.2033. Epub 2011 Mar 13.
Qijun Chen, Liliya A Yatsunyk, John M Nicoludis, Mark S Garcia, Ramon Kranaster, Shankar Balasubramanian, David Monchaud, Marie-Paule Teulade-Fichou, Lara Abramowitz, David C Schultz, F Brad Johnson
Affiliations
- PMID: 21399640
- PMCID: PMC3119813
- DOI: 10.1038/nsmb.2033
Rudimentary G-quadruplex-based telomere capping in Saccharomyces cerevisiae
Jasmine S Smith et al. Nat Struct Mol Biol. 2011 Apr.
Abstract
Telomere capping conceals chromosome ends from exonucleases and checkpoints, but the full range of capping mechanisms is not well defined. Telomeres have the potential to form G-quadruplex (G4) DNA, although evidence for telomere G4 DNA function in vivo is limited. In budding yeast, capping requires the Cdc13 protein and is lost at nonpermissive temperatures in cdc13-1 mutants. Here, we use several independent G4 DNA-stabilizing treatments to suppress cdc13-1 capping defects. These include overexpression of three different G4 DNA binding proteins, loss of the G4 DNA unwinding helicase Sgs1, or treatment with small molecule G4 DNA ligands. In vitro, we show that protein-bound G4 DNA at a 3' overhang inhibits 5'→3' resection of a paired strand by exonuclease I. These findings demonstrate that, at least in the absence of full natural capping, G4 DNA can play a positive role at telomeres in vivo.
Figures
Figure 1
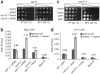
Overexpression of the G4 DNA binding protein Stm1 rescues growth defects caused by telomere uncapping and is independent of _RAD52_-dependent homologous recombination. (a) Growth of cdc13-1 mutants carrying pSTM1 or vector alone at permissive temperature (PT, 22 °C) or at semipermissive temperature (SPT, 30 °C). (b) pSTM1 overexpression rescues impaired growth caused by telomere uncapping in stn1-154 mutants at SPT. (c) Growth of cdc13-1 rad52Δ mutants carrying pSTM1 or vector. For each assay, serial dilutions of cells were spotted on selective medium and grown for 3 d (b,c) or 4 d (a). (d) Top: map of a typical telomere containing two tandem subtelomeric Y’ elements, separated by interstitial telomere repeats. Bottom: telomere Southern blots of samples grown in liquid culture at SPT for 2 d. Type I and type II survivors of telomerase inactivation are shown for comparison. The different sizes of internal Y’ fragments are due to short and long forms of Y’. Lanes 1–5 and 6–9 are sections from the same Southern blot.
Figure 2
Expression of two additional, distinct G4 DNA binding proteins rescues the cdc13-1 temperature-sensitive phenotype. (a) The G4 DNA-binding Sgs1 RQC domain, overexpressed from a plasmid and under the control of the GAL1 promoter (pRQC), or vector control were transformed into the indicated strain backgrounds and tested in spot assays. (b) A 5× HA-tagged Sgs1 RQC domain is enriched at telomeres of cdc13-1 mutants. The primer sets used were directly adjacent to the telomeres (subtelomeric Y’ repeat) or a control, centromere-proximal primer set (to a portion of SWC4, which has no QFP). The telomeric/centromeric ratio is listed above bars. Error bars are ± 1 s.d., and essentially the same result was obtained in three independent ChIP experiments. Mock IP indicates that cdc13-1 sgs1Δ cells with the RQC-expressing vector were ChIPed without antibody. (c) GAL-induced overexpression of the G4 DNA binding single-chain antibody (scFv), HF1 or empty vector in cdc13-1 mutants. For each assay, serial dilutions of cells were spotted on selective medium containing 2% galactose and grown at the indicated temperatures for 4 d. (d) The 13xMyc-tagged HF1 scFv is enriched at telomeres of cdc13-1 mutants. DNA was amplified as described in (b). Error bars are ± 1 s.d., and essentially the same result was obtained in three independent ChIP experiments.
Figure 3
Loss of the SGS1 activities associated with G4 DNA binding and unwinding rescues cdc13-1 temperature sensitivity and is independent of _RAD52_-dependent homologous recombination. (a) Growth of cdc13-1 rad24Δ rad52Δ, either SGS1 or sgs1Δ, at permissive temperature versus SPT. (b) Map of specific Sgs1 domains mutated or deleted in this study. (c) Deletion of the G-quadruplex binding RQC domain alone is sufficient to rescue the cdc13-1 growth defect. (d) Loss of Sgs1 helicase activity (K706A point mutation, denoted sgs1-hd) is sufficient to rescue cdc13-1 temperature sensitivity. (e) Sgs1 is the sole component of the Sgs1–Top3–Rmi1 complex that, when lost, is sufficient to rescue the cdc13-1 temperature sensitive phenotype. For each assay, serial dilutions of cells were spotted on YPAD (a,e) or selective medium (c,d) and grown at the indicated temperatures for 3 d (a,c,d) or 4 d (e).
Figure 4
Telomere-proximal single-stranded (ss) DNA accumulation at NPT (37 °C) is attenuated by two G4 DNA-stabilizing mechanisms. (a,b) ssDNA measurements in the context of STM1 overexpression (a) or sgs1 deletion (b) in cdc13-1 rad24Δ rad52Δ backgrounds. Subtelomeric G-strand ssDNA was probed with a complementary end-labeled ssDNA probe against the Y’ element and quantified by normalizing hybridization signals of native to denatured samples. Each sample was spotted in triplicate, and standard errors are shown. Each graph is one representative experiment. The apparent difference in the timing of ssDNA accumulation between the experiments reflects the time points examined rather than true experimental variability. (c,d) Attenuation of Rad53 phosphorylation by Stm1 overexpression or sgs1 deletion in cdc13-1 rad24Δ rad52Δ cells incubated at NPT (37 °C). Treatment of wild-type cells with 0.033% methyl methanesulfonate (MMS) provides a positive control for Rad53 phosphorylation (4d, lane 1).
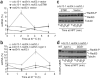
Figure 5
Diminished rescue of cdc13-1 growth at SPT by sgs1 deletion in cells with mutant telomerase RNA templates that decrease QFP at telomeres. (a) cdc13-1 rad24Δ rad52Δ tlc1Δ cells, either SGS1 or sgs1Δ, and with plasmid-borne wild-type TLC1 or the uCu or CuA mutant tlc1 alleles, were spotted on SC-HIS medium and grown for 4 d. The sequence of the TLC1 template, including the mutated region (bold), is as follows: 3′-CACACACCCACACCAC-5′. (b) CD spectra of representative tlc1 mutant telomere sequences and their corrected counterparts (see Table 1). (c) Thermal difference spectra (TDS) of mutant versus corrected sequences, shown as the difference between molar extinction coefficients. G4 DNA yields a negative peak at 295 nm and positive peaks at 273 and 242 nm. All four corrected sequences showed evidence of G4 DNA formation by means of CD (parallel quadruplex for 1-corr and mixed parallel and antiparallel for 2, 3 and 4-corr) and TDS. (d) Thermal denaturation of oligonucleotides was followed by CD absorbance at 263 nm (for oligonucleotides 1, 1-corrected (corr), 2 and 2-corr) or 290 nm (for oligonucleotides 3, 3-corr, 4 and 4-corr).
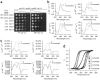
Figure 6
G4 DNA-binding proteins and G4 DNA-forming sequences cooperate to inhibit end-resection by Exo1 in vitro. (a) Synthetic substrates having 3′ overhangs that form (bottom) or do not form (top) G-quadruplex DNA. Each contains 51 bp of identical duplex DNA. The 5′ end of each top strand was biotinylated (B) to inhibit resection from the incorrect end, and the terminal base of each bottom strand was labeled with α-32P-dCTP. We illustrate a parallel intramolecular quadruplex (bottom), although this may not be the actual or only structure formed by this overhang. (b) Analysis of overhang substrates by CD shows a mixture of parallel and anti-parallel G4 DNA (GGG overhang) and a possible GA homoduplex (GAG overhang) (see Supplementary Fig. 6) under the exact conditions used for the Exo1 assay. (c) Thermal difference spectra of GGG and GAG yield a G-quadruplex-specific signal for only the GGG overhang. (d,e) Both Stm1 (d) and HF1 (e) cooperate with G4 DNA overhang to partially rescue Exo1 5′→3′ resection. Quantification of the fraction of remaining full-length sequence; shown are the averages of two (d) or three (e) independent experiments with standard errors and _P_-values from unpaired two-tailed Student’s _t_-tests. (f) Quantification of protection by T4 gene 32 protein (T4g32p). An average of two experiments is shown. P > 0.7 for all concentrations of T4g32p.
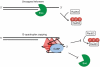
Figure 7
Model of telomere capping in which G4 DNA cooperates with G-quadruplex binding proteins (G4BP) to inhibit exonucleolytic degradation and checkpoint activation caused by loss of Cdc13 function.
Similar articles
- Survival and growth of yeast without telomere capping by Cdc13 in the absence of Sgs1, Exo1, and Rad9.
Ngo HP, Lydall D. Ngo HP, et al. PLoS Genet. 2010 Aug 19;6(8):e1001072. doi: 10.1371/journal.pgen.1001072. PLoS Genet. 2010. PMID: 20808892 Free PMC article. - Linear chromosome maintenance in the absence of essential telomere-capping proteins.
Zubko MK, Lydall D. Zubko MK, et al. Nat Cell Biol. 2006 Jul;8(7):734-40. doi: 10.1038/ncb1428. Epub 2006 Jun 11. Nat Cell Biol. 2006. PMID: 16767084 - Role of folding kinetics of secondary structures in telomeric G-overhangs in the regulation of telomere maintenance in Saccharomyces cerevisiae.
Jurikova K, Gajarsky M, Hajikazemi M, Nosek J, Prochazkova K, Paeschke K, Trantirek L, Tomaska L. Jurikova K, et al. J Biol Chem. 2020 Jul 3;295(27):8958-8971. doi: 10.1074/jbc.RA120.012914. Epub 2020 May 8. J Biol Chem. 2020. PMID: 32385108 Free PMC article. - Cell cycle regulation of G-quadruplex DNA structures at telomeres.
Juranek SA, Paeschke K. Juranek SA, et al. Curr Pharm Des. 2012;18(14):1867-72. doi: 10.2174/138161212799958404. Curr Pharm Des. 2012. PMID: 22376109 Review. - G-Quadruplexes at Telomeres: Friend or Foe?
Bryan TM. Bryan TM. Molecules. 2020 Aug 13;25(16):3686. doi: 10.3390/molecules25163686. Molecules. 2020. PMID: 32823549 Free PMC article. Review.
Cited by
- The evolving world of protein-G-quadruplex recognition: a medicinal chemist's perspective.
Sissi C, Gatto B, Palumbo M. Sissi C, et al. Biochimie. 2011 Aug;93(8):1219-30. doi: 10.1016/j.biochi.2011.04.018. Epub 2011 Apr 29. Biochimie. 2011. PMID: 21549174 Free PMC article. Review. - Markers of cellular senescence. Telomere shortening as a marker of cellular senescence.
Bernadotte A, Mikhelson VM, Spivak IM. Bernadotte A, et al. Aging (Albany NY). 2016 Jan;8(1):3-11. doi: 10.18632/aging.100871. Aging (Albany NY). 2016. PMID: 26805432 Free PMC article. Review. - Hypothesis: Paralog Formation from Progenitor Proteins and Paralog Mutagenesis Spur the Rapid Evolution of Telomere Binding Proteins.
Lustig AJ. Lustig AJ. Front Genet. 2016 Feb 10;7:10. doi: 10.3389/fgene.2016.00010. eCollection 2016. Front Genet. 2016. PMID: 26904098 Free PMC article. - DNA repair at telomeres: keeping the ends intact.
Webb CJ, Wu Y, Zakian VA. Webb CJ, et al. Cold Spring Harb Perspect Biol. 2013 Jun 1;5(6):a012666. doi: 10.1101/cshperspect.a012666. Cold Spring Harb Perspect Biol. 2013. PMID: 23732473 Free PMC article. Review. - Repetitive DNA loci and their modulation by the non-canonical nucleic acid structures R-loops and G-quadruplexes.
Hall AC, Ostrowski LA, Pietrobon V, Mekhail K. Hall AC, et al. Nucleus. 2017 Mar 4;8(2):162-181. doi: 10.1080/19491034.2017.1292193. Nucleus. 2017. PMID: 28406751 Free PMC article. Review.
References
- Palm W, de Lange T. How shelterin protects mammalian telomeres. Annu. Rev. Genet. 2008;42:301–334. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- T32 GM07229/GM/NIGMS NIH HHS/United States
- P01 AG031862-03/AG/NIA NIH HHS/United States
- A5709/CRUK_/Cancer Research UK/United Kingdom
- T32 GM007229/GM/NIGMS NIH HHS/United States
- T32 GM008216-22/GM/NIGMS NIH HHS/United States
- P01 AG031862/AG/NIA NIH HHS/United States
- T32 GM008216/GM/NIGMS NIH HHS/United States
- P01 AG031862-02/AG/NIA NIH HHS/United States
- R01 AG021521/AG/NIA NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases