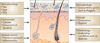The skin microbiome - PubMed (original) (raw)
Review
The skin microbiome
Elizabeth A Grice et al. Nat Rev Microbiol. 2011 Apr.
Erratum in
- Nat Rev Microbiol. 2011 Aug;9(8):626
Abstract
The skin is the human body's largest organ, colonized by a diverse milieu of microorganisms, most of which are harmless or even beneficial to their host. Colonization is driven by the ecology of the skin surface, which is highly variable depending on topographical location, endogenous host factors and exogenous environmental factors. The cutaneous innate and adaptive immune responses can modulate the skin microbiota, but the microbiota also functions in educating the immune system. The development of molecular methods to identify microorganisms has led to an emerging view of the resident skin bacteria as highly diverse and variable. An enhanced understanding of the skin microbiome is necessary to gain insight into microbial involvement in human skin disorders and to enable novel promicrobial and antimicrobial therapeutic approaches for their treatment.
Figures
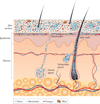
Figure 1. Schematic of skin histology viewed in cross-section with microorganisms and skin appendages
Microorganisms (viruses, bacteria and fungi) and mites cover the surface of the skin and reside deep in the hair and glands. On the skin surface, rod and round bacteria — such as Proteobacteria and Staphylococcus spp., respectively — form communities that are deeply intertwined among themselves and other microorganisms. Commensal fungi such as Malassezia spp. grow both as branching filamentous hypha and as individual cells. Virus particles live both freely and in bacterial cells. Skin mites, such as Demodex folliculorum and Demodex brevis, are some of the smallest arthropods and live in or near hair follicles. Skin appendages include hair follicles, sebaceous glands and sweat glands.
Figure 2. Factors contributing to variation in the skin microbiome
Exogenous and endogenous factors discussed in this Review that contribute to variation between individuals and over the lifetime of an individual.
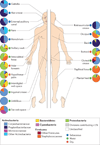
Figure 3. Topographical distribution of bacteria on skin sites
The skin microbiome is highly dependent on the microenvironment of the sampled site. The family-level classification of bacteria colonizing an individual subject is shown, with the phyla in bold. The sites selected were those that show a predilection for skin bacterial infections and are grouped as sebaceous or oily (blue circles), moist (typically skin creases) (green circles) and dry, flat surfaces (red circles). The sebaceous sites are: glabella (between the eyebrows); alar crease (side of the nostril); external auditory canal (inside the ear); retroauricular crease (behind the ear); occiput (back of the scalp); manubrium (upper chest); and back. Moist sites are: nare (inside the nostril); axillary vault (armpit); antecubital fossa (inner elbow); interdigital web space (between the middle and ring fingers); inguinal crease (side of the groin); gluteal crease (topmost part of the fold between the buttocks); popliteal fossa (behind the knee); plantar heel (bottom of the heel of the foot); toe web space; and umbilicus (navel). Dry sites are: volar forearm (inside of the mid-forearm); hypothenar palm (palm of the hand proximal to the little finger); and buttock. Data from REF. 42.
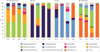
Figure 4. Interpersonal variation of the skin microbiome
The microbial distribution of four sites on four healthy volunteers (HV1, HV2, HV3 and HV4) is depicted at the antecubital fold (inner elbow; part a); the back (part b); the nare (inside the nostril; part c); and the plantar heel (bottom of the heel of the foot; part d). Skin microbial variation is more dependent on the site than on the individual. Bars represent the relative abundance of bacterial taxa as determined by 16S ribosomal RNA sequencing. Data from REF. 42.
Similar articles
- The skin microbiome: current perspectives and future challenges.
Chen YE, Tsao H. Chen YE, et al. J Am Acad Dermatol. 2013 Jul;69(1):143-55. doi: 10.1016/j.jaad.2013.01.016. Epub 2013 Mar 13. J Am Acad Dermatol. 2013. PMID: 23489584 Free PMC article. Review. - Functions of the skin microbiota in health and disease.
Sanford JA, Gallo RL. Sanford JA, et al. Semin Immunol. 2013 Nov 30;25(5):370-7. doi: 10.1016/j.smim.2013.09.005. Epub 2013 Nov 20. Semin Immunol. 2013. PMID: 24268438 Free PMC article. Review. - The microbiome in patients with atopic dermatitis.
Paller AS, Kong HH, Seed P, Naik S, Scharschmidt TC, Gallo RL, Luger T, Irvine AD. Paller AS, et al. J Allergy Clin Immunol. 2019 Jan;143(1):26-35. doi: 10.1016/j.jaci.2018.11.015. Epub 2018 Nov 23. J Allergy Clin Immunol. 2019. PMID: 30476499 Free PMC article. Review. - Commensal bacteria and cutaneous immunity.
Nakamizo S, Egawa G, Honda T, Nakajima S, Belkaid Y, Kabashima K. Nakamizo S, et al. Semin Immunopathol. 2015 Jan;37(1):73-80. doi: 10.1007/s00281-014-0452-6. Epub 2014 Oct 18. Semin Immunopathol. 2015. PMID: 25326105 Review. - Microbiology of the skin: resident flora, ecology, infection.
Roth RR, James WD. Roth RR, et al. J Am Acad Dermatol. 1989 Mar;20(3):367-90. doi: 10.1016/s0190-9622(89)70048-7. J Am Acad Dermatol. 1989. PMID: 2645319 Review.
Cited by
- Induced Transient Immune Tolerance in Ticks and Vertebrate Host: A Keystone of Tick-Borne Diseases?
Boulanger N, Wikel S. Boulanger N, et al. Front Immunol. 2021 Feb 12;12:625993. doi: 10.3389/fimmu.2021.625993. eCollection 2021. Front Immunol. 2021. PMID: 33643313 Free PMC article. Review. - Fermentation of Propionibacterium acnes, a commensal bacterium in the human skin microbiome, as skin probiotics against methicillin-resistant Staphylococcus aureus.
Shu M, Wang Y, Yu J, Kuo S, Coda A, Jiang Y, Gallo RL, Huang CM. Shu M, et al. PLoS One. 2013;8(2):e55380. doi: 10.1371/journal.pone.0055380. Epub 2013 Feb 6. PLoS One. 2013. PMID: 23405142 Free PMC article. - Valorisation of tomato pomace in anti-pollution and microbiome-balance face cream.
Rajkowska K, Otlewska A, Raczyk A, Maciejczyk E, Krajewska A. Rajkowska K, et al. Sci Rep. 2024 Sep 3;14(1):20516. doi: 10.1038/s41598-024-71323-4. Sci Rep. 2024. PMID: 39227423 Free PMC article. - Epithelium Expressing the E7 Oncoprotein of HPV16 Attracts Immune-Modulatory Dendritic Cells to the Skin and Suppresses Their Antigen-Processing Capacity.
Chandra J, Miao Y, Romoff N, Frazer IH. Chandra J, et al. PLoS One. 2016 Mar 31;11(3):e0152886. doi: 10.1371/journal.pone.0152886. eCollection 2016. PLoS One. 2016. PMID: 27031095 Free PMC article. - The facial microbiome and metabolome across different geographic regions.
Tao R, Li T, Wang Y, Wang R, Li R, Bianchi P, Duplan H, Zhang Y, Li H, Wang R. Tao R, et al. Microbiol Spectr. 2024 Jan 11;12(1):e0324823. doi: 10.1128/spectrum.03248-23. Epub 2023 Dec 8. Microbiol Spectr. 2024. PMID: 38063390 Free PMC article.
References
- Chiller K, Selkin BA, Murakawa GJ. Skin microflora and bacterial infections of the skin. J. Investig. Dermatol. Symp. Proc. 2001;6:170–174. - PubMed
- Fredricks DN. Microbial ecology of human skin in health and disease. J. Investig. Dermatol. Symp. Proc. 2001;6:167–169. - PubMed
- Marples M. The Ecology of the Human Skin. Bannerstone House, Springfield, Illinois: Charles C Thomas; 1965. A seminal and comprehensive work of classical dermatological microbiology.
- Roth RR, James WD. Microbial ecology of the skin. Annu. Rev. Microbiol. 1988;42:441–464. - PubMed
- Noble WC. Skin microbiology: coming of age. J. Med. Microbiol. 1984;17:1–12. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources