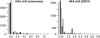Partial short-read sequencing of a highly inbred Iberian pig and genomics inference thereof - PubMed (original) (raw)
Partial short-read sequencing of a highly inbred Iberian pig and genomics inference thereof
A Esteve-Codina et al. Heredity (Edinb). 2011 Sep.
Abstract
Despite dramatic reduction in sequencing costs with the advent of next generation sequencing technologies, obtaining a complete mammalian genome sequence at sufficient depth is still costly. An alternative is partial sequencing. Here, we have sequenced a reduced representation library of an Iberian sow from the Guadyerbas strain, a highly inbred strain that has been used in numerous QTL studies because of its extreme phenotypic characteristics. Using the Illumina Genome Analyzer II (San Diego, CA, USA), we resequenced ∼ 1% of the genome with average 4 × depth, identifying 68,778 polymorphisms. Of these, 55,457 were putative fixed differences with respect to the assembly, based on the genome of a Duroc pig, and 13,321 were heterozygous positions within Guadyerbas. Despite being highly inbred, the estimate of heterozygosity within Guadyerbas was ∼ 0.78 kb(-1) in autosomes, after correcting for low depth. Nucleotide variability was consistently higher at the telomeric regions than on the rest of the chromosome, likely a result of increased recombination rates. Further, variability was 50% lower in the X-chromosome than in autosomes, which may be explained by a recent bottleneck or by selection. We divided the whole genome in 500 kb windows and we analyzed overrepresented gene ontology terms in regions of low and high variability. Multi organism process, pigmentation and cell killing were overrepresented in high variability regions and metabolic process ontology, within low variability regions. Further, a genome wide Hudson-Kreitman-Aguadé test was carried out per window; overall, variability was in agreement with neutral expectations.
Figures
Figure 1
Simulated isolation with migration model that represents the Iberian/Duroc history (the public assembly pertains to a Duroc sow). The Duroc and Iberian populations descend from an ancestral population harboring a nucleotide diversity θ_=4_Neμ; after the split τ generations ago, both breeds of effective sizes N DU and N IB may have interchanged individuals with rate m. A mixed coalescence and gene dropping procedure was used.
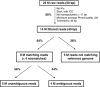
Figure 2
Bioinformatics pipeline.
Figure 3
Lowess adjusted curves of variability in chromosomes 4, 7, 14 and X. An increased variability is observed towards the telomeres in metacentric chromosomes 4 and X, whereas the ratio is distorted in SSC7 because of high SLA variability near window 50; SSC14 is acrocentric. Solid red line, Iberian heterozygosity (ĥ); dashed black line, Iberian—Duroc heterozygosity (f̂). Position refers to window number. A full color version of this figure is available at the Heredity journal online.
Figure 4
Histograms comparing observed (black bars) and simulated (grey bars) HKA statistics across autosomal and sex chromosome windows. The simulated results correspond to parameter values that minimized the Wilcoxon statistics.
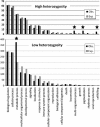
Figure 5
Expected and observed gene ontology counts among genes located in high and low variability windows. Bars with asterisk are significant (P<0.001) overrepresented gene ontologies.
Similar articles
- Dissecting structural and nucleotide genome-wide variation in inbred Iberian pigs.
Esteve-Codina A, Paudel Y, Ferretti L, Raineri E, Megens HJ, Silió L, Rodríguez MC, Groenen MA, Ramos-Onsins SE, Pérez-Enciso M. Esteve-Codina A, et al. BMC Genomics. 2013 Mar 5;14:148. doi: 10.1186/1471-2164-14-148. BMC Genomics. 2013. PMID: 23497037 Free PMC article. - Comprehensive inbred variation discovery in Bama pigs using de novo assemblies.
Zhang L, Huang Y, Si J, Wu Y, Wang M, Jiang Q, Guo Y, Liang J, Lan G. Zhang L, et al. Gene. 2018 Dec 30;679:81-89. doi: 10.1016/j.gene.2018.08.051. Epub 2018 Sep 1. Gene. 2018. PMID: 30179680 - Application of massive parallel sequencing to whole genome SNP discovery in the porcine genome.
Amaral AJ, Megens HJ, Kerstens HH, Heuven HC, Dibbits B, Crooijmans RP, den Dunnen JT, Groenen MA. Amaral AJ, et al. BMC Genomics. 2009 Aug 12;10:374. doi: 10.1186/1471-2164-10-374. BMC Genomics. 2009. PMID: 19674453 Free PMC article. - Bioinformatics approaches for genomics and post genomics applications of next-generation sequencing.
Horner DS, Pavesi G, Castrignanò T, De Meo PD, Liuni S, Sammeth M, Picardi E, Pesole G. Horner DS, et al. Brief Bioinform. 2010 Mar;11(2):181-97. doi: 10.1093/bib/bbp046. Epub 2009 Oct 27. Brief Bioinform. 2010. PMID: 19864250 Review. - Understanding genetic variation and function- the applications of next generation sequencing.
Harrison RJ. Harrison RJ. Semin Cell Dev Biol. 2012 Apr;23(2):230-6. doi: 10.1016/j.semcdb.2012.01.006. Epub 2012 Jan 20. Semin Cell Dev Biol. 2012. PMID: 22285423 Review.
Cited by
- Maternal Transmission Ratio Distortion in Two Iberian Pig Varieties.
Vázquez-Gómez M, Hijas-Villalba MM, Varona L, Ibañez-Escriche N, Rosas JP, Negro S, Noguera JL, Casellas J. Vázquez-Gómez M, et al. Genes (Basel). 2020 Sep 5;11(9):1050. doi: 10.3390/genes11091050. Genes (Basel). 2020. PMID: 32899475 Free PMC article. - Whole genome SNPs discovery in Nero Siciliano pig.
D'Alessandro E, Giosa D, Sapienza I, Giuffrè L, Cigliano RA, Romeo O, Zumbo A. D'Alessandro E, et al. Genet Mol Biol. 2019 Jul-Sep;42(3):594-602. doi: 10.1590/1678-4685-GMB-2018-0169. Epub 2019 Nov 14. Genet Mol Biol. 2019. PMID: 31188930 Free PMC article. - Inbreeding depression load for litter size in Entrepelado and Retinto Iberian pig varieties1.
Casellas J, Ibáñez-Escriche N, Varona L, Rosas JP, Noguera JL. Casellas J, et al. J Anim Sci. 2019 Apr 29;97(5):1979-1986. doi: 10.1093/jas/skz084. J Anim Sci. 2019. PMID: 30869129 Free PMC article. - Contrasting Patterns of Genomic Diversity Reveal Accelerated Genetic Drift but Reduced Directional Selection on X-Chromosome in Wild and Domestic Sheep Species.
Chen ZH, Zhang M, Lv FH, Ren X, Li WR, Liu MJ, Nam K, Bruford MW, Li MH. Chen ZH, et al. Genome Biol Evol. 2018 Apr 1;10(5):1282-1297. doi: 10.1093/gbe/evy085. Genome Biol Evol. 2018. PMID: 29790980 Free PMC article. - Genome-wide genetic structure and differentially selected regions among Landrace, Erhualian, and Meishan pigs using specific-locus amplified fragment sequencing.
Li Z, Wei S, Li H, Wu K, Cai Z, Li D, Wei W, Li Q, Chen J, Liu H, Zhang L. Li Z, et al. Sci Rep. 2017 Aug 30;7(1):10063. doi: 10.1038/s41598-017-09969-6. Sci Rep. 2017. PMID: 28855565 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources