Comprehensive identification of protein substrates of the Dot/Icm type IV transporter of Legionella pneumophila - PubMed (original) (raw)
Comprehensive identification of protein substrates of the Dot/Icm type IV transporter of Legionella pneumophila
Wenhan Zhu et al. PLoS One. 2011.
Abstract
A large number of proteins transferred by the Legionella pneumophila Dot/Icm system have been identified by various strategies. With no exceptions, these strategies are based on one or more characteristics associated with the tested proteins. Given the high level of diversity exhibited by the identified proteins, it is possible that some substrates have been missed in these screenings. In this study, we took a systematic method to survey the L. pneumophila genome by testing hypothetical orfs larger than 300 base pairs for Dot/Icm-dependent translocation. 798 of the 832 analyzed orfs were successfully fused to the carboxyl end of β-lactamase. The transfer of the fusions into mammalian cells was determined using the β-lactamase reporter substrate CCF4-AM. These efforts led to the identification of 164 proteins positive in translocation. Among these, 70 proteins are novel substrates of the Dot/Icm system. These results brought the total number of experimentally confirmed Dot/Icm substrates to 275. Sequence analysis of the C-termini of these identified proteins revealed that Lpg2844, which contains few features known to be important for Dot/Icm-dependent protein transfer can be translocated at a high efficiency. Thus, our efforts have identified a large number of novel substrates of the Dot/Icm system and have revealed the diverse features recognizable by this protein transporter.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
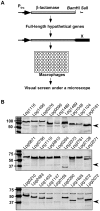
Figure 1. Construction of a library expressing fusions of β-lactamase and L. pneumophila hypothetical proteins.
A. A schematic structure of the fusion proteins and the screening strategy. In most cases, the gene was fused with the β-lactamase by inserting into the vector as a _Bam_HI/_Sal_I fragment. After infection, samples were loaded with the CCF4-AM dye and were inspected under a fluorescence microscope. B. Evaluation of the library for expression of the fusion proteins. Plasmids directing expression of β-lactamase fusions were introduced into a wild type L. pneumophila strain. Total cell lysates of bacteria grown in the presence of IPTG were used to examine the steady state of the fusion proteins by immunoblot. In each image, the detection of a protein nonspecifically recognized by the antibody (arrow) was used as a loading control.
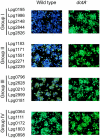
Figure 2. Dot/Icm-dependent translocation of substrates.
Effectors identified in this study were divided into four groups according to their transfer efficiencies, 5 genes from each group were shown as representatives. U937 cells seeded in 96-well plates were infected with wild type or _dot/icm_-deficient L. pneumophila strains expressing a gene fusion and images were acquired 2 hrs after CCF4-AM loading with a DP72 color camera (Olympus). Group I, genes with translocation efficiency >90%; Group II, genes with translocation efficiency between 50% and 80%; Group III, genes with translocation efficiency between 20% and 45% and Group IV, genes with translocation efficiency less than 15%.
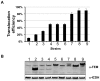
Figure 3. Translocation efficiency does not correlate with levels of β-lactamase fusion expressed in L. pneumophila.
A. The translocation efficiency of 9 substrates in the β-lactamase assay. After CCF4-AM loading, macrophages infected with bacterial strains expression fusions between β-lactamase and individual genes were inspected under a fluorescence microscope, translocation efficiencies were obtained by enumerating cells emitting blue and green fluorescence signals, respectively. Experiments were performed in triplicates and at least 300 cells were counted each sample. Similar results were obtained in at least two independent experiments. B. The levels of the fusion proteins in L. pneumophila strains used for infections shown in A. Bacterial cells equivalent to one OD600 unit were lysed in 200 µl of SDS loading buffer, 15 µl of boiled supernatant were resolved by SDS-PAGE. After transferring to nitrocellulose membranes, the fusion protein was detected with a β-lactamase specific antibody by immunoblot. The isocitrate dehydrogenase (ICDH) was probed as a loading control. Samples: 1. Lpg1776; 2, Lpg0021; 3, Lpg2425; 4, Lpg1147; 5, Lpg0181; 6, Lpg2555; 7, Lpg2874; 8, Lpg0405; 9, Lpg0195.
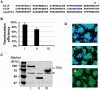
Figure 4. Diverse features presented in the C-terminal end of Dot/Icm substrates.
A. Alignment of the last 50 amino acids of three well-established effectors and the new substrate Lpg2844 to highlight the features important for translocation found in Dot/Icm substrates, including: i) The hydrophobic residue at the -3rd position (▽, red) and ii) the E-block (the three blue residues in SidF). Note the different amino acids composition in Lpg2844. B–D. A region containing the last 100 amino acids of Lpg2844 is important and sufficient for promoting translocation. Bacterial strains expressing fusions of β-lactamase to full-length Lpg2844 (I), it's last 100 aa (II) or a fragment lacking the last 100 aa (III) were used to infect macrophages and infected cells were loaded with the CCF4-AM dye. Translocation efficiency (B) was obtained as described in Fig. 3, data shown are the average of three independent experiments done in triplicates; stable expression of the fusions by L. pneumophila, equal amount of protein samples resolved by SDS-PAGE was probed for the fusions with a β-lactamase specific antibody. The ca. 60-kDa non-specific band detected by the antibody was used as a loading control (arrow in panel C). Representative images of infected cells loaded with CCF4-AM (D). Similar results were obtained in at least two independent experiments.
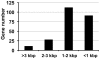
Figure 5. The distribution of all verified Dot/Icm substrates according to their length.
Proteins experimentally shown as substrates of the Dot/Icm transporter were collected and sorted according to the length of the gene, which were then divided into four groups: I, genes larger than 3 kbp; II, genes ranging between 2 to 3 kbp; III, genes ranging between 1 to 2 kbp and IV, genes smaller than 1 kbp.
Similar articles
- The E Block motif is associated with Legionella pneumophila translocated substrates.
Huang L, Boyd D, Amyot WM, Hempstead AD, Luo ZQ, O'Connor TJ, Chen C, Machner M, Montminy T, Isberg RR. Huang L, et al. Cell Microbiol. 2011 Feb;13(2):227-45. doi: 10.1111/j.1462-5822.2010.01531.x. Epub 2010 Nov 3. Cell Microbiol. 2011. PMID: 20880356 Free PMC article. - A C-terminal translocation signal required for Dot/Icm-dependent delivery of the Legionella RalF protein to host cells.
Nagai H, Cambronne ED, Kagan JC, Amor JC, Kahn RA, Roy CR. Nagai H, et al. Proc Natl Acad Sci U S A. 2005 Jan 18;102(3):826-31. doi: 10.1073/pnas.0406239101. Epub 2004 Dec 21. Proc Natl Acad Sci U S A. 2005. PMID: 15613486 Free PMC article. - Multiple substrates of the Legionella pneumophila Dot/Icm system identified by interbacterial protein transfer.
Luo ZQ, Isberg RR. Luo ZQ, et al. Proc Natl Acad Sci U S A. 2004 Jan 20;101(3):841-6. doi: 10.1073/pnas.0304916101. Epub 2004 Jan 8. Proc Natl Acad Sci U S A. 2004. PMID: 14715899 Free PMC article. - Effector translocation by the Legionella Dot/Icm type IV secretion system.
Qiu J, Luo ZQ. Qiu J, et al. Curr Top Microbiol Immunol. 2013;376:103-15. doi: 10.1007/82_2013_345. Curr Top Microbiol Immunol. 2013. PMID: 23918176 Review. - Legionella pneumophila Dot/Icm translocated substrates: a sum of parts.
Ensminger AW, Isberg RR. Ensminger AW, et al. Curr Opin Microbiol. 2009 Feb;12(1):67-73. doi: 10.1016/j.mib.2008.12.004. Epub 2009 Jan 20. Curr Opin Microbiol. 2009. PMID: 19157961 Free PMC article. Review.
Cited by
- Implication of the VirD4 coupling protein of the Lvh type 4 secretion system in virulence phenotypes of Legionella pneumophila.
Bandyopadhyay P, Lang EA, Rasaputra KS, Steinman HM. Bandyopadhyay P, et al. J Bacteriol. 2013 Aug;195(15):3468-75. doi: 10.1128/JB.00430-13. Epub 2013 May 31. J Bacteriol. 2013. PMID: 23729650 Free PMC article. - Reassessing the role of DotF in the Legionella pneumophila type IV secretion system.
Sutherland MC, Binder KA, Cualing PY, Vogel JP. Sutherland MC, et al. PLoS One. 2013 Jun 7;8(6):e65529. doi: 10.1371/journal.pone.0065529. Print 2013. PLoS One. 2013. PMID: 23762385 Free PMC article. - Legionella longbeachae effector protein RavZ inhibits autophagy and regulates phagosome ubiquitination during infection.
Shi Y, Liu H, Ma K, Luo ZQ, Qiu J. Shi Y, et al. PLoS One. 2023 Feb 9;18(2):e0281587. doi: 10.1371/journal.pone.0281587. eCollection 2023. PLoS One. 2023. PMID: 36758031 Free PMC article. - The Influence of _Acanthamoeba_-Legionella Interaction in the Virulence of Two Different Legionella Species.
Gomes TS, Gjiknuri J, Magnet A, Vaccaro L, Ollero D, Izquierdo F, Fenoy S, Hurtado C, Del Águila C. Gomes TS, et al. Front Microbiol. 2018 Dec 5;9:2962. doi: 10.3389/fmicb.2018.02962. eCollection 2018. Front Microbiol. 2018. PMID: 30568639 Free PMC article. - A Legionella pneumophila amylase is essential for intracellular replication in human macrophages and amoebae.
Best A, Price C, Ozanic M, Santic M, Jones S, Abu Kwaik Y. Best A, et al. Sci Rep. 2018 Apr 20;8(1):6340. doi: 10.1038/s41598-018-24724-1. Sci Rep. 2018. PMID: 29679057 Free PMC article.
References
- Swanson MS, Hammer BK. Legionella pneumophila pathogesesis: a fateful journey from amoebae to macrophages. Annu Rev Microbiol. 2000;54:567–613. - PubMed
- Vogel JP, Andrews HL, Wong SK, Isberg RR. Conjugative transfer by the virulence system of Legionella pneumophila. . Science. 1998;279:873–876. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01AI069344/AI/NIAID NIH HHS/United States
- R01 AI069344/AI/NIAID NIH HHS/United States
- K02AI085403/AI/NIAID NIH HHS/United States
- R21 AI092043/AI/NIAID NIH HHS/United States
- R21AI092043/AI/NIAID NIH HHS/United States
- K02 AI085403/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources