Incorporating distant sequence features and radial basis function networks to identify ubiquitin conjugation sites - PubMed (original) (raw)
Incorporating distant sequence features and radial basis function networks to identify ubiquitin conjugation sites
Tzong-Yi Lee et al. PLoS One. 2011.
Abstract
Ubiquitin (Ub) is a small protein that consists of 76 amino acids about 8.5 kDa. In ubiquitin conjugation, the ubiquitin is majorly conjugated on the lysine residue of protein by Ub-ligating (E3) enzymes. Three major enzymes participate in ubiquitin conjugation. They are E1, E2 and E3 which are responsible for activating, conjugating and ligating ubiquitin, respectively. Ubiquitin conjugation in eukaryotes is an important mechanism of the proteasome-mediated degradation of a protein and regulating the activity of transcription factors. Motivated by the importance of ubiquitin conjugation in biological processes, this investigation develops a method, UbSite, which uses utilizes an efficient radial basis function (RBF) network to identify protein ubiquitin conjugation (ubiquitylation) sites. This work not only investigates the amino acid composition but also the structural characteristics, physicochemical properties, and evolutionary information of amino acids around ubiquitylation (Ub) sites. With reference to the pathway of ubiquitin conjugation, the substrate sites for E3 recognition, which are distant from ubiquitylation sites, are investigated. The measurement of F-score in a large window size (-20∼+20) revealed a statistically significant amino acid composition and position-specific scoring matrix (evolutionary information), which are mainly located distant from Ub sites. The distant information can be used effectively to differentiate Ub sites from non-Ub sites. As determined by five-fold cross-validation, the model that was trained using the combination of amino acid composition and evolutionary information performs best in identifying ubiquitin conjugation sites. The prediction sensitivity, specificity, and accuracy are 65.5%, 74.8%, and 74.5%, respectively. Although the amino acid sequences around the ubiquitin conjugation sites do not contain conserved motifs, the cross-validation result indicates that the integration of distant sequence features of Ub sites can improve predictive performance. Additionally, the independent test demonstrates that the proposed method can outperform other ubiquitylation prediction tools.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
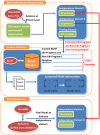
Figure 1. The analytic flowchart of UbSite.
Figure 2. The detailed process of generating position specific scoring matrix (PSSM) and encoding the fragment of amino acid sequence by generated PSSM.
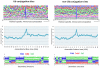
Figure 3. The position-specific amino acid composition, accessible surface area and secondary structure of ubiquitin conjugated lysines and non-ubiquitin conjugated lysines.
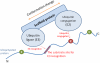
Figure 4. The hypothetic model of identifying the distant sequence features for E3 recognition.
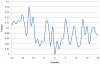
Figure 5. The statistically significant composition of amino acids for each position in the window length from −20 to +20.
Based on the measurement of F-score, the positions −16, −10, −3, −1, +1, +5, +10, +13, and +17, containing higher value of F-score, are significant for differentiating the ubiquitylation sites from non-ubiquitylation sites.
Figure 6. The statistically significant evolutionary information of amino acids for each position in the window length from −20 to +20.
Based on the measurement of F-score, the positions −19, −17, −15, −12, −10, −4, −1, +5, +9, +13, +15 and +18, containing higher value of F-score, are significant for differentiating the ubiquitylation sites from non-ubiquitylation sites.
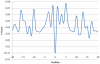
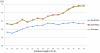
Figure 7. The predictive performance of the models trained with different window length varying from 11-mer to 41-mer.
Similar articles
- UbiSite: incorporating two-layered machine learning method with substrate motifs to predict ubiquitin-conjugation site on lysines.
Huang CH, Su MG, Kao HJ, Jhong JH, Weng SL, Lee TY. Huang CH, et al. BMC Syst Biol. 2016 Jan 11;10 Suppl 1(Suppl 1):6. doi: 10.1186/s12918-015-0246-z. BMC Syst Biol. 2016. PMID: 26818456 Free PMC article. - Characterization and identification of ubiquitin conjugation sites with E3 ligase recognition specificities.
Nguyen VN, Huang KY, Huang CH, Chang TH, Bretaña N, Lai K, Weng J, Lee TY. Nguyen VN, et al. BMC Bioinformatics. 2015;16 Suppl 1(Suppl 1):S1. doi: 10.1186/1471-2105-16-S1-S1. Epub 2015 Jan 21. BMC Bioinformatics. 2015. PMID: 25707307 Free PMC article. - Proteome-wide identification of ubiquitylation sites by conjugation of engineered lysine-less ubiquitin.
Oshikawa K, Matsumoto M, Oyamada K, Nakayama KI. Oshikawa K, et al. J Proteome Res. 2012 Feb 3;11(2):796-807. doi: 10.1021/pr200668y. Epub 2011 Nov 23. J Proteome Res. 2012. PMID: 22053931 - The Ubiquitin Code in Disease Pathogenesis and Progression: Composition, Characteristics and its Potential as a Therapeutic Target.
Lee JS, Kim HY, Kwon YT, Ji CH, Lee SJ, Kim SB. Lee JS, et al. Discov Med. 2025 Feb;37(193):203-221. doi: 10.24976/Discov.Med.202537193.18. Discov Med. 2025. PMID: 39973547 Review. - Structural basis of generic versus specific E2-RING E3 interactions in protein ubiquitination.
Gundogdu M, Walden H. Gundogdu M, et al. Protein Sci. 2019 Oct;28(10):1758-1770. doi: 10.1002/pro.3690. Epub 2019 Aug 23. Protein Sci. 2019. PMID: 31340062 Free PMC article. Review.
Cited by
- Optimization of auto-induction medium for G-CSF production by Escherichia coli using artificial neural networks coupled with genetic algorithm.
Tian H, Liu C, Gao XD, Yao WB. Tian H, et al. World J Microbiol Biotechnol. 2013 Mar;29(3):505-13. doi: 10.1007/s11274-012-1204-1. Epub 2012 Nov 7. World J Microbiol Biotechnol. 2013. PMID: 23132252 - Characterization and Identification of Lysine Succinylation Sites based on Deep Learning Method.
Huang KY, Hsu JB, Lee TY. Huang KY, et al. Sci Rep. 2019 Nov 7;9(1):16175. doi: 10.1038/s41598-019-52552-4. Sci Rep. 2019. PMID: 31700141 Free PMC article. - Investigation and identification of protein carbonylation sites based on position-specific amino acid composition and physicochemical features.
Weng SL, Huang KY, Kaunang FJ, Huang CH, Kao HJ, Chang TH, Wang HY, Lu JJ, Lee TY. Weng SL, et al. BMC Bioinformatics. 2017 Mar 14;18(Suppl 3):66. doi: 10.1186/s12859-017-1472-8. BMC Bioinformatics. 2017. PMID: 28361707 Free PMC article. - Characterization and identification of lysine crotonylation sites based on machine learning method on both plant and mammalian.
Wang R, Wang Z, Wang H, Pang Y, Lee TY. Wang R, et al. Sci Rep. 2020 Nov 24;10(1):20447. doi: 10.1038/s41598-020-77173-0. Sci Rep. 2020. PMID: 33235255 Free PMC article. - Investigation and identification of protein γ-glutamyl carboxylation sites.
Lee TY, Lu CT, Chen SA, Bretaña NA, Cheng TH, Su MG, Huang KY. Lee TY, et al. BMC Bioinformatics. 2011;12 Suppl 13(Suppl 13):S10. doi: 10.1186/1471-2105-12-S13-S10. Epub 2011 Nov 30. BMC Bioinformatics. 2011. PMID: 22372765 Free PMC article.
References
- Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. - PubMed
- Ou CY, Pi H, Chien CT. Control of protein degradation by E3 ubiquitin ligases in Drosophila eye development. Trends Genet. 2003;19:382–389. - PubMed
- Hicke L, Schubert HL, Hill CP. Ubiquitin-binding domains. Nat Rev Mol Cell Biol. 2005;6:610–621. - PubMed
- Peng J, Schwartz D, Elias JE, Thoreen CC, Cheng D, et al. A proteomics approach to understanding protein ubiquitination. Nat Biotechnol. 2003;21:921–926. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials