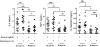Human macrophage responses to clinical isolates from the Mycobacterium tuberculosis complex discriminate between ancient and modern lineages - PubMed (original) (raw)
Human macrophage responses to clinical isolates from the Mycobacterium tuberculosis complex discriminate between ancient and modern lineages
Damien Portevin et al. PLoS Pathog. 2011 Mar.
Abstract
The aim of the present study was to determine whether there is a correlation between phylogenetic relationship and inflammatory response amongst a panel of clinical isolates representative of the global diversity of the human Mycobacterium tuberculosis Complex (MTBC). Measurement of cytokines from infected human peripheral blood monocyte-derived macrophages revealed a wide variation in the response to different strains. The same pattern of high or low response to individual strains was observed for different pro-inflammatory cytokines and chemokines, and was conserved across multiple human donors. Although each major phylogenetic lineage of MTBC included strains inducing a range of cytokine responses, we found that overall inflammatory phenotypes differed significantly across lineages. In particular, comparison of evolutionarily modern lineages demonstrated a significant skewing towards lower early inflammatory response. The differential response to ancient and modern lineages observed using GM-CSF derived macrophages was also observed in autologous monocyte-derived dendritic cells and murine bone marrow-derived macrophages, but not in human unfractionated peripheral blood mononuclear cells. We hypothesize that the reduced immune responses to modern lineages contribute to more rapid disease progression and transmission, which might be a selective advantage in the context of expanding human populations. In addition to the lineage effects, the large strain-to-strain variation in innate immune responses elicited by MTBC will need to be considered in tuberculosis vaccine development.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
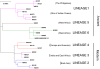
Figure 1. Selection of MTBC isolates representative of global genetic diversity.
Phylogenetic tree of the 26 strains used for the study based on a concatenate alignment of 89 genes previously described . One strain belonging to Lineage 3 and three strains to Lineage 6 were not present in the original study and are depicted with dashed lines branching from the node representing the most common recent ancestor of the respective lineage. The color code highlights the six lineages of M. tuberculosis complex (MTBC) strains affecting humans. These lineages were clustered in two groups according to the occurrence of the TbD1 genomic deletion which discriminates “ancient” from “modern” lineages , .
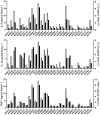
Figure 2. M. tuberculosis complex clinical isolates vary widely in their induction of pro-inflammatory cytokines.
Graphic representation of the cytokine response from GM-CSF monocyte-derived macrophages of two different donors after infection with the panel of MTBC strains. IL-6, IL-12p40/p70 and TNFα were measured by ELISA from filtered supernatants 24 h post-infection (MOI 1∶1). The inflammatory response differs widely between the different strains. Cytokine responses from each donor were plotted on different scales to highlight the conservation of the trend between donors.
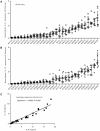
Figure 3. Strain-related hierarchy in pro-inflammatory response is maintained across multiple human donors.
A) Scatter plot representation of IL-6 and B) IL-12p40/p70 macrophage responses from eight different individual donors. Cytokine responses from each individual donor to a particular strain were normalized with respect to the median response of the donor towards the panel of strains. The response to each strain was ranked according to the average response across the eight donors. The results show that the hierarchy of IL-6 and IL-12p40/p70 response is maintained across independent donors. C) Linear regression analysis shows significant correlation between IL-6 and IL-12p40/p70 macrophage responses to the different strains used in the study. Correlation coefficient and p-value result from non-parametric correlation test (Spearman) are indicated on the graph.
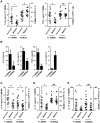
Figure 4. IL-6 production varies with genetic clustering of MTBC lineages.
A) Median normalized levels of cytokine production by human monocyte-derived macrophages from eight independent donors were averaged for scatter plot representation and clustered according to strain lineage. Non-parametric Kruskal-Wallis test revealed a significant effect of lineage variable in cytokine production (P<0.001). B) PCA plot (PC1 versus PC3) of the strains used in this study based on the polymorphisms found between them in Hershberg et al. 2008 highlights three main groups clearly separating the three modern lineages from Lineage 1, and from the two M. africanum lineages (5 and 6). C) IL-6 response from T1-MDMs from eight donors infected by the panel of MTBC strains indicates that Lineages 5+6 and Lineage 1 induce more IL-6 than the modern lineages. (Mann-Whitney U test, * P<0.05, ** P<0.01).
Figure 5. Strains from the modern lineages of M. tuberculosis complex induce lower levels of pro-inflammatory cytokines and chemokines.
Supernatants of infected macrophages 24 h post-infection from 3 independent donors were pooled and analyzed using multiplex technology and results were plotted for each individual cytokine/chemokine. The results showed statistically significant lower responses by macrophages infected with strains belonging to the modern lineage (Mann-Whitney U test, * P<0.05, ** P<0.01).
Figure 6. Lower cytokines production during infection with modern strains is associated with a delayed inflammatory response.
Scatter plot representation of macrophage cytokine responses to ancient versus modern strains at 24 h and 72 h post-infection from a single donor. Supernatants from infected macrophages were collected at 24 h and 72 h and analyzed by ELISA for IL-6 and IL-12p40/p70 content. While the trend towards higher cytokine production from macrophages infected by strains belonging to the ancient lineage is maintained, convergence of cytokine levels at the later time point (particularly in the case of IL-6) suggests that lower responses to the modern lineage may reflect a delay rather than complete inhibition of the inflammatory response (Mann-Whitney U test, * P<0.05, NS non significant).
Figure 7. The inflammatory response to ancient and modern lineages differs when infecting autologous PBMCs or anti-inflammatory Type 2 macrophages.
A) Scatter plot representation of macrophage (circles) versus autologous PBMCs (squares) cytokine responses to ancient (open) versus modern (filled) groups of strains shows that the pro-inflammatory phenotype was reversed when infecting PBMCs instead of GM-CSF derived macrophages from the same donor and looking at IL-6 production but not TNFα. B) Macrophages from three independent donors were derived from monocytes either in the presence of recombinant human GM-CSF or M-CSF to generate Type 1 macrophages (T1-MDMs) and Type 2 macrophages (T2-MDMs) respectively. Results are presented as histograms of cytokine production from LPS-stimulated T1-MDMs and T2-MDMs showing increased pro-inflammatory potential of T1-MDMs, revealed by higher levels of IL-6, IL-12p40/p70 and TNFα produced 24 h post-stimulation, in contrast to T2-MDMs which produced higher levels of anti-inflammatory IL-10. C) IL-6 D) IL-12p40/p70 E) IL-10 averaged levels from autologous T1-MDMs versus T2-MDMs (single donor) infected by the various strains of MTBC clustered into ancient and modern lineages. The higher pro-inflammatory phenotype associated with the ancient lineages is only observed for IL-6 when infecting T2-MDMs but not for IL-12p40/p70 neither for the production of anti-inflammatory IL-10 (Mann-Whitney U test, * P<0.05, ** P<0.01, NS non significant).
Figure 8. The delayed inflammatory response of modern lineage is not due to IL-10.
Scatter plot representation of Type 1 macrophage cytokine response from a single donor 24 h post-infection towards ancient (circles) versus modern (squares) groups of strains in the presence (filled) or absence (open) of anti-IL-10 blocking antibodies. IL-10 blockage resulted in a significant increase in the production of IL-6 and TNFα independent of lineage. The delayed inflammatory response to strains from modern lineages is not due to IL-10 regulatory activity (Paired t-test for intra-group comparison and Mann-Whitney U test for inter-group comparison, * P<0.05, ** P<0.01 and *** P<0.001).
Figure 9. The difference in inflammatory response to ancient and modern lineages is also observed in monocyte-derived DCs although the pattern of secreted cytokines/chemokines is partially different.
Scatter plot representation of monocyte derived dendritic cell cytokine responses towards ancient versus modern groups of strains. Filtered supernatants of MD-DCs 24 h post-infection from three independent donors were pooled, analyzed using multiplex technology and resulting values were plotted for each individual cytokine/chemokine. Results show a significant trend towards higher production of each cytokine or chemokine from MD-DCs infected by strains belonging to the ancient lineages (Mann-Whitney U test, * P<0.05, **P<0.01).
Similar articles
- Clinical isolates of the modern Mycobacterium tuberculosis lineage 4 evade host defense in human macrophages through eluding IL-1β-induced autophagy.
Romagnoli A, Petruccioli E, Palucci I, Camassa S, Carata E, Petrone L, Mariano S, Sali M, Dini L, Girardi E, Delogu G, Goletti D, Fimia GM. Romagnoli A, et al. Cell Death Dis. 2018 May 24;9(6):624. doi: 10.1038/s41419-018-0640-8. Cell Death Dis. 2018. PMID: 29795378 Free PMC article. - Shaping the niche in macrophages: Genetic diversity of the M. tuberculosis complex and its consequences for the infected host.
Reiling N, Homolka S, Kohl TA, Steinhäuser C, Kolbe K, Schütze S, Brandenburg J. Reiling N, et al. Int J Med Microbiol. 2018 Jan;308(1):118-128. doi: 10.1016/j.ijmm.2017.09.009. Epub 2017 Sep 14. Int J Med Microbiol. 2018. PMID: 28969988 Review. - Bacterial diversity dominates variable macrophage responses of tuberculosis patients in Tanzania.
Hiza H, Zwyer M, Hella J, Arbués A, Sasamalo M, Borrell S, Xu ZM, Ross A, Brites D, Fellay J, Reither K, Gagneux S, Portevin D. Hiza H, et al. Sci Rep. 2024 Apr 23;14(1):9287. doi: 10.1038/s41598-024-60001-0. Sci Rep. 2024. PMID: 38653771 Free PMC article. - Clade-specific virulence patterns of Mycobacterium tuberculosis complex strains in human primary macrophages and aerogenically infected mice.
Reiling N, Homolka S, Walter K, Brandenburg J, Niwinski L, Ernst M, Herzmann C, Lange C, Diel R, Ehlers S, Niemann S. Reiling N, et al. mBio. 2013 Jul 30;4(4):e00250-13. doi: 10.1128/mBio.00250-13. mBio. 2013. PMID: 23900170 Free PMC article. - Host-pathogen coevolution in human tuberculosis.
Gagneux S. Gagneux S. Philos Trans R Soc Lond B Biol Sci. 2012 Mar 19;367(1590):850-9. doi: 10.1098/rstb.2011.0316. Philos Trans R Soc Lond B Biol Sci. 2012. PMID: 22312052 Free PMC article. Review.
Cited by
- Two new rapid SNP-typing methods for classifying Mycobacterium tuberculosis complex into the main phylogenetic lineages.
Stucki D, Malla B, Hostettler S, Huna T, Feldmann J, Yeboah-Manu D, Borrell S, Fenner L, Comas I, Coscollà M, Gagneux S. Stucki D, et al. PLoS One. 2012;7(7):e41253. doi: 10.1371/journal.pone.0041253. Epub 2012 Jul 20. PLoS One. 2012. PMID: 22911768 Free PMC article. - Characterisation of secretome-based immune responses of human leukocytes infected with various Mycobacterium tuberculosis lineages.
Kaewseekhao B, Roytrakul S, Yingchutrakul Y, Laohaviroj M, Salao K, Faksri K. Kaewseekhao B, et al. PeerJ. 2021 Jun 3;9:e11565. doi: 10.7717/peerj.11565. eCollection 2021. PeerJ. 2021. PMID: 34141493 Free PMC article. - Global Distribution and Evolution of Mycobacterium bovis Lineages.
Zimpel CK, Patané JSL, Guedes ACP, de Souza RF, Silva-Pereira TT, Camargo NCS, de Souza Filho AF, Ikuta CY, Neto JSF, Setubal JC, Heinemann MB, Guimaraes AMS. Zimpel CK, et al. Front Microbiol. 2020 May 7;11:843. doi: 10.3389/fmicb.2020.00843. eCollection 2020. Front Microbiol. 2020. PMID: 32477295 Free PMC article. - Mycobacterium tuberculosis cyclophilin A uses novel signal sequence for secretion and mimics eukaryotic cyclophilins for interaction with host protein repertoire.
Bhaduri A, Misra R, Maji A, Bhetaria PJ, Mishra S, Arora G, Singh LK, Dhasmana N, Dubey N, Virdi JS, Singh Y. Bhaduri A, et al. PLoS One. 2014 Feb 4;9(2):e88090. doi: 10.1371/journal.pone.0088090. eCollection 2014. PLoS One. 2014. PMID: 24505389 Free PMC article. - Mapping of genotype-phenotype diversity among clinical isolates of mycobacterium tuberculosis by sequence-based transcriptional profiling.
Rose G, Cortes T, Comas I, Coscolla M, Gagneux S, Young DB. Rose G, et al. Genome Biol Evol. 2013;5(10):1849-62. doi: 10.1093/gbe/evt138. Genome Biol Evol. 2013. PMID: 24115728 Free PMC article.
References
- Gagneux S, Small PM. Global phylogeography of Mycobacterium tuberculosis and implications for tuberculosis product development. Lancet Infect Dis. 2007;7:328–337. - PubMed
- Nicol MP, Wilkinson RJ. The clinical consequences of strain diversity in Mycobacterium tuberculosis. Trans R Soc Trop Med Hyg. 2008;102:955–965. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- MC_U117581288/MRC_/Medical Research Council/United Kingdom
- MC_U117588500/MRC_/Medical Research Council/United Kingdom
- U117588500/MRC_/Medical Research Council/United Kingdom
- U117581288/MRC_/Medical Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Medical