A clarification of the terms used in comparing semi-automated particle selection algorithms in cryo-EM - PubMed (original) (raw)
A clarification of the terms used in comparing semi-automated particle selection algorithms in cryo-EM
Robert Langlois et al. J Struct Biol. 2011 Sep.
Erratum in
- J Struct Biol. 2015 Mar;189(3):281
Abstract
Many cyro-EM datasets are heterogeneous stemming from molecules undergoing conformational changes. The need to characterize each of the substrates with sufficient resolution entails a large increase in the data flow and motivates the development of more effective automated particle selection algorithms. Concepts and procedures from the machine-learning field are increasingly employed toward this end. However, a review of recent literature has revealed a discrepancy in terminology of the performance scores used to compare particle selection algorithms, and this has subsequently led to ambiguities in the meaning of claimed performance. In an attempt to curtail the perpetuation of this confusion and to disentangle past mistakes, we review the performance of published particle selection efforts with a set of explicitly defined performance scores using the terminology established and accepted within the field of machine learning.
Copyright © 2011 Elsevier Inc. All rights reserved.
Figures
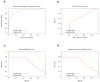
Figure 1
Metric curves comparing two classifiers on the Keyhole Limpet Hemocyanin dataset, a publicly available dataset used in the 2004 particle selection bakeoff (Zhu et al., 2004). The title “false discovery curve” (b) was coined for lack of a better term. Note that the curve shown in (d) depicts a proper linear interpolation, rather than the non-linear interpolation, which would produce a smooth curve similar to a ROC plot.
Similar articles
- Reference-free particle selection enhanced with semi-supervised machine learning for cryo-electron microscopy.
Langlois R, Pallesen J, Frank J. Langlois R, et al. J Struct Biol. 2011 Sep;175(3):353-61. doi: 10.1016/j.jsb.2011.06.004. Epub 2011 Jun 17. J Struct Biol. 2011. PMID: 21708269 Free PMC article. - Automatic post-picking using MAPPOS improves particle image detection from cryo-EM micrographs.
Norousi R, Wickles S, Leidig C, Becker T, Schmid VJ, Beckmann R, Tresch A. Norousi R, et al. J Struct Biol. 2013 May;182(2):59-66. doi: 10.1016/j.jsb.2013.02.008. Epub 2013 Feb 21. J Struct Biol. 2013. PMID: 23454482 - AutoCryoPicker: an unsupervised learning approach for fully automated single particle picking in Cryo-EM images.
Al-Azzawi A, Ouadou A, Tanner JJ, Cheng J. Al-Azzawi A, et al. BMC Bioinformatics. 2019 Jun 13;20(1):326. doi: 10.1186/s12859-019-2926-y. BMC Bioinformatics. 2019. PMID: 31195977 Free PMC article. - [A review of automatic particle recognition in Cryo-EM images].
Wu X, Wu X. Wu X, et al. Sheng Wu Yi Xue Gong Cheng Xue Za Zhi. 2010 Oct;27(5):1178-82. Sheng Wu Yi Xue Gong Cheng Xue Za Zhi. 2010. PMID: 21089695 Review. Chinese. - Automated Modeling and Validation of Protein Complexes in Cryo-EM Maps.
Cragnolini T, Sweeney A, Topf M. Cragnolini T, et al. Methods Mol Biol. 2021;2215:189-223. doi: 10.1007/978-1-0716-0966-8_9. Methods Mol Biol. 2021. PMID: 33368005 Review.
Cited by
- Automated particle picking for low-contrast macromolecules in cryo-electron microscopy.
Langlois R, Pallesen J, Ash JT, Nam Ho D, Rubinstein JL, Frank J. Langlois R, et al. J Struct Biol. 2014 Apr;186(1):1-7. doi: 10.1016/j.jsb.2014.03.001. Epub 2014 Mar 6. J Struct Biol. 2014. PMID: 24607413 Free PMC article. - DeepCryoPicker: fully automated deep neural network for single protein particle picking in cryo-EM.
Al-Azzawi A, Ouadou A, Max H, Duan Y, Tanner JJ, Cheng J. Al-Azzawi A, et al. BMC Bioinformatics. 2020 Nov 9;21(1):509. doi: 10.1186/s12859-020-03809-7. BMC Bioinformatics. 2020. PMID: 33167860 Free PMC article. - Semi-automated selection of cryo-EM particles in RELION-1.3.
Scheres SH. Scheres SH. J Struct Biol. 2015 Feb;189(2):114-22. doi: 10.1016/j.jsb.2014.11.010. Epub 2014 Dec 6. J Struct Biol. 2015. PMID: 25486611 Free PMC article. - Automatic cryo-EM particle selection for membrane proteins in spherical liposomes.
Liu Y, Sigworth FJ. Liu Y, et al. J Struct Biol. 2014 Mar;185(3):295-302. doi: 10.1016/j.jsb.2014.01.004. Epub 2014 Jan 24. J Struct Biol. 2014. PMID: 24468290 Free PMC article. - Reference-free particle selection enhanced with semi-supervised machine learning for cryo-electron microscopy.
Langlois R, Pallesen J, Frank J. Langlois R, et al. J Struct Biol. 2011 Sep;175(3):353-61. doi: 10.1016/j.jsb.2011.06.004. Epub 2011 Jun 17. J Struct Biol. 2011. PMID: 21708269 Free PMC article.
References
- Adiga U, Baxter WT, Hall RJ, Rockel B, Rath BK, et al. Particle picking by segmentation: A comparative study with SPIDER-based manual particle picking. Journal of Structural Biology. 2005;152:211–220. - PubMed
- Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society Series B (Methodological) 1995;57:289–300.
- Brodersen KH, Ong CS, Stephan KE, Buhmann JM. The Binormal Assumption on Precision-Recall Curves. Pattern Recognition (ICPR); 20th International Conference on; 2010. pp. 4263–4266.pp. 4263–4266.
- Caruana R, Niculescu-Mizil A. An empirical comparison of supervised learning algorithms. Proceedings of the 23rd international conference on Machine learning; ACM, Pittsburgh, Pennsylvania. 2006.
- Chen JZ, Grigorieff N. SIGNATURE: A single-particle selection system for molecular electron microscopy. Journal of Structural Biology. 2007;157:168–173. - PubMed
Publication types
MeSH terms
Grants and funding
- R37 GM029169/GM/NIGMS NIH HHS/United States
- R37 GM 29169/GM/NIGMS NIH HHS/United States
- R37 GM029169-25/GM/NIGMS NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- R01 GM055440/GM/NIGMS NIH HHS/United States
- R01 GM 55440/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Miscellaneous