Assessing and improving methods used in operational taxonomic unit-based approaches for 16S rRNA gene sequence analysis - PubMed (original) (raw)
Assessing and improving methods used in operational taxonomic unit-based approaches for 16S rRNA gene sequence analysis
Patrick D Schloss et al. Appl Environ Microbiol. 2011 May.
Abstract
In spite of technical advances that have provided increases in orders of magnitude in sequencing coverage, microbial ecologists still grapple with how to interpret the genetic diversity represented by the 16S rRNA gene. Two widely used approaches put sequences into bins based on either their similarity to reference sequences (i.e., phylotyping) or their similarity to other sequences in the community (i.e., operational taxonomic units [OTUs]). In the present study, we investigate three issues related to the interpretation and implementation of OTU-based methods. First, we confirm the conventional wisdom that it is impossible to create an accurate distance-based threshold for defining taxonomic levels and instead advocate for a consensus-based method of classifying OTUs. Second, using a taxonomic-independent approach, we show that the average neighbor clustering algorithm produces more robust OTUs than other hierarchical and heuristic clustering algorithms. Third, we demonstrate several steps to reduce the computational burden of forming OTUs without sacrificing the robustness of the OTU assignment. Finally, by blending these solutions, we propose a new heuristic that has a minimal effect on the robustness of OTUs and significantly reduces the necessary time and memory requirements. The ability to quickly and accurately assign sequences to OTUs and then obtain taxonomic information for those OTUs will greatly improve OTU-based analyses and overcome many of the challenges encountered with phylotype-based methods.
Figures
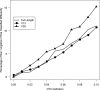
Fig. 1.
Cumulative fraction of taxa that had a specified maximum intrataxon distance (A) and total branch length (B) for each taxonomic level when full-length 16S rRNA gene sequences were analyzed. At each taxonomic level, sequences that did not affiliate with a known lineage (i.e., incertae sedis) were excluded. The numbers in parentheses next to the name of each taxonomic level indicate the number of taxa within that level that we observed. (See Fig. S1 and S2 in the supplemental material for the same analysis using the V13 and V35 sequences, respectively.)
Fig. 2.
Fraction of OTUs calculated for a 0.03-cutoff level that were represented by more than one sequence and had different classifications when we classified the OTU using a representative sequence from the OTU or by determining the majority consensus taxonomy for the full-length, V13, and V35 16S rRNA gene sequence data sets.
Fig. 3.
Variation in the Matthew's correlation coefficient calculated for OTUs identified by using eight classification algorithms at genetic distances varying between 0.00 and 0.10 for full-length 16S rRNA gene sequences. (See Fig. S3 and S4 in the supplemental material for the same analysis using the V13 and V35 sequences, respectively.)
Fig. 4.
Comparison of the Matthew's correlation coefficients for OTUs calculated from a threshold of 0.00 to 0.10 when using the phylotype-OTU heuristic for full-length 16S rRNA gene sequences. For each region, cutoff, and taxonomic level used to split the sequences, the correlation coefficients overlapped with each other, except for the family and genus taxonomic levels. (See Fig. S5 and S6 in the supplemental material for the same analysis using the V13 and V35 sequences, respectively.)
References
- Baldi P., Brunak S., Chauvin Y., Andersen C. A., Nielsen H. 2000. Assessing the accuracy of prediction algorithms for classification: an overview. Bioinformatics 16:412–424 - PubMed
- Cohan F. M. 2002. What are bacterial species? Annu. Rev. Microbiol. 56:457–487 - PubMed
- Edgar R. C. 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources