A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression - PubMed (original) (raw)
. 2011 Apr 7;472(7341):120-4.
doi: 10.1038/nature09819. Epub 2011 Mar 20.
Yul W Yang, Bo Liu, Amartya Sanyal, Ryan Corces-Zimmerman, Yong Chen, Bryan R Lajoie, Angeline Protacio, Ryan A Flynn, Rajnish A Gupta, Joanna Wysocka, Ming Lei, Job Dekker, Jill A Helms, Howard Y Chang
Affiliations
- PMID: 21423168
- PMCID: PMC3670758
- DOI: 10.1038/nature09819
A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression
Kevin C Wang et al. Nature. 2011.
Abstract
The genome is extensively transcribed into long intergenic noncoding RNAs (lincRNAs), many of which are implicated in gene silencing. Potential roles of lincRNAs in gene activation are much less understood. Development and homeostasis require coordinate regulation of neighbouring genes through a process termed locus control. Some locus control elements and enhancers transcribe lincRNAs, hinting at possible roles in long-range control. In vertebrates, 39 Hox genes, encoding homeodomain transcription factors critical for positional identity, are clustered in four chromosomal loci; the Hox genes are expressed in nested anterior-posterior and proximal-distal patterns colinear with their genomic position from 3' to 5'of the cluster. Here we identify HOTTIP, a lincRNA transcribed from the 5' tip of the HOXA locus that coordinates the activation of several 5' HOXA genes in vivo. Chromosomal looping brings HOTTIP into close proximity to its target genes. HOTTIP RNA binds the adaptor protein WDR5 directly and targets WDR5/MLL complexes across HOXA, driving histone H3 lysine 4 trimethylation and gene transcription. Induced proximity is necessary and sufficient for HOTTIP RNA activation of its target genes. Thus, by serving as key intermediates that transmit information from higher order chromosomal looping into chromatin modifications, lincRNAs may organize chromatin domains to coordinate long-range gene activation.
©2011 Macmillan Publishers Limited. All rights reserved
Figures
Figure 1. HOTTIP is a lincRNA transcribed in distal anatomic sites
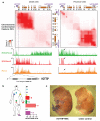
(A) Chromatin state map of distal vs. proximal cells. Top panels: Chromosome Conformation Capture-Carbon Copy (5C) analysis of distal (foreskin) and proximal (lung) human fibroblasts. Heat map representations (generated by my5C) of 5C data (bin size 30 kb, step size 3 kb) for HOXA in foreskin and lung fibroblasts. Red intensity of each pixel indicates relative interaction between the two points on the genomic coordinates. The diagonal represents frequent cis interactions between regions located in close proximity along the linear genome. 5C signals that are away from the diagonal represent long-range looping interactions. Bottom: Chromatin occupancy across HOXA. X-axis is genomic coordinate; Y-axis depicts occupancy of the indicated histone marks or protein (ChIP/input). Box and arrows highlight chromatin states of HOTTIP. (B) HOTTIP expression in primary human fibroblasts from 11 anatomic sites. Mean + S.E. is shown in all panels with error bars. (C) In situ hybridization of HOTTIP in E13.5 mouse embryo.
Figure 2. HOTTIP is required for coordinate activation of 5′ HOXA genes
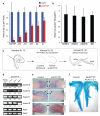
(A) Knockdown of HOTTIP abrogates expression of 5′ HOXA genes in foreskin fibroblasts, but not HOXD genes or BID (B). Mean ± s.d. are shown. (C) Schematic of chick RNAi experiment. (D) HOTTIP is required for 5′ HoxA gene expression in vivo. RT-PCR of the indicated genes from control or HOTTIP-depleted distal limb bud is shown; quantitation and normalization by Actin signal is shown below each band. GAG signal confirms successful retroviral transduction in all cases. (E) In situ hybridization of 5′ HoxA genes in chick limb buds. Arrowheads highlight distal domains of high HoxA gene expression that are impacted by HOTTIP knockdown. (F) Shortening of distal bony elements in HOTTIP-depleted forelimbs. Alcian blue staining highlights the skeletal elements. Red and purple lines highlight radius and 3rd digit lengths, respectively.
Figure 3. HOTTIP is required for the active chromatin state of 5′ HOXA cluster
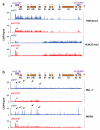
(A) Knockdown of HOTTIP broadly decreases H3K4me3 across 5′ HOXA locus but focally affects H3K27me3 at HOTTIP itself. Display is as in Fig. 1A. (B) Knockdown of HOTTIP abrogates peaks of MLL1 and WDR5 occupancy near TSSs of 5′ HOXA genes and leads to accumulation of these proteins at HOTTIP itself. Arrows highlight peaks of MLL1 and WDR5 occupancy; open arrowheads highlight chromatin state of HOTTIP upon HOTTIP RNA knockdown.
Figure 4. HOTTIP programs active chromatin via WDR5
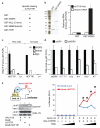
(A) Summary of RNA-protein interaction studies. Each of the indicated recombinant protein was purified and used to retrieve purified HOTTIP or control histone RNA in vitro. Only GST-WDR5 specifically retrieved HOTTIP. (B) HOTTIP binds directly and specifically to WDR5. Left: Purified GST and GST-WDR5 are visualized by SDS-PAGE and Comassie Blue staining. Right: Retrieved RNAs are quantified by qRT-PCR. (C) HOTTIP binds specifically to WDR5 in cells. IP of endogenous WDR5 protein from PC3 (prostate) and T24 (bladder) carcinoma cells specifically retrieved HOTTIP, but not control IPs with IgG or chromatin binder SIRT6. (D) WDR5 is required for 5′ HOXA gene expression, including HOTTIP. (E) HOTTIP recruitment potentiates transcription. Left: The BoxB tethering system. BoxB-RNA specifically binds λN fused to GAL4 DNA binding domain, recruiting the complex to a UAS-luciferase reporter gene. After transient transfection, IP of GAL4- λN specifically retrieves BoxB-HOTTIP. Right: Luciferase activity after co-transfection of the indicated constructs (mean ± s.d., _n_=4 triplicates, * indciates p<0.05 Student’s _t_-test comparing BoxB-LacZ vs. BoxB-HOTTIP). Sloped triangle indicates increasing input of plasmids encoding ncRNAs.
Comment in
- Non-coding RNA: HOTTIP goes the distance.
Burgess DJ. Burgess DJ. Nat Rev Genet. 2011 May;12(5):300. doi: 10.1038/nrg2992. Epub 2011 Apr 12. Nat Rev Genet. 2011. PMID: 21483457 No abstract available.
Similar articles
- Long noncoding RNA HOTTIP cooperates with CCCTC-binding factor to coordinate HOXA gene expression.
Wang F, Tang Z, Shao H, Guo J, Tan T, Dong Y, Lin L. Wang F, et al. Biochem Biophys Res Commun. 2018 Jun 12;500(4):852-859. doi: 10.1016/j.bbrc.2018.04.173. Epub 2018 May 1. Biochem Biophys Res Commun. 2018. PMID: 29698677 - Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs.
Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, Chang HY. Rinn JL, et al. Cell. 2007 Jun 29;129(7):1311-23. doi: 10.1016/j.cell.2007.05.022. Cell. 2007. PMID: 17604720 Free PMC article. - SET1A/COMPASS and shadow enhancers in the regulation of homeotic gene expression.
Cao K, Collings CK, Marshall SA, Morgan MA, Rendleman EJ, Wang L, Sze CC, Sun T, Bartom ET, Shilatifard A. Cao K, et al. Genes Dev. 2017 Apr 15;31(8):787-801. doi: 10.1101/gad.294744.116. Epub 2017 May 9. Genes Dev. 2017. PMID: 28487406 Free PMC article. - The HOTTIP (HOXA transcript at the distal tip) lncRNA: Review of oncogenic roles in human.
Ghafouri-Fard S, Dashti S, Taheri M. Ghafouri-Fard S, et al. Biomed Pharmacother. 2020 Jul;127:110158. doi: 10.1016/j.biopha.2020.110158. Epub 2020 Apr 23. Biomed Pharmacother. 2020. PMID: 32335298 Review. - Molecular leveraging of HOX-embedded non-coding RNAs in the progression of acute myeloid leukemia.
Wilson C, Swaroop P, Kumar S, Chopra A, Sharawat SK. Wilson C, et al. Hum Cell. 2024 Nov 30;38(1):24. doi: 10.1007/s13577-024-01149-9. Hum Cell. 2024. PMID: 39614990 Review.
Cited by
- lincRNAs: genomics, evolution, and mechanisms.
Ulitsky I, Bartel DP. Ulitsky I, et al. Cell. 2013 Jul 3;154(1):26-46. doi: 10.1016/j.cell.2013.06.020. Cell. 2013. PMID: 23827673 Free PMC article. Review. - Long noncoding RNAs in cardiac development and ageing.
Devaux Y, Zangrando J, Schroen B, Creemers EE, Pedrazzini T, Chang CP, Dorn GW 2nd, Thum T, Heymans S; Cardiolinc network. Devaux Y, et al. Nat Rev Cardiol. 2015 Jul;12(7):415-25. doi: 10.1038/nrcardio.2015.55. Epub 2015 Apr 7. Nat Rev Cardiol. 2015. PMID: 25855606 Review. - HCC-Related lncRNAs: Roles and Mechanisms.
Shah M, Sarkar D. Shah M, et al. Int J Mol Sci. 2024 Jan 2;25(1):597. doi: 10.3390/ijms25010597. Int J Mol Sci. 2024. PMID: 38203767 Free PMC article. Review. - The context of gene expression regulation.
Gibcus JH, Dekker J. Gibcus JH, et al. F1000 Biol Rep. 2012;4:8. doi: 10.3410/B4-8. Epub 2012 Apr 2. F1000 Biol Rep. 2012. PMID: 22500194 Free PMC article. - Long non-coding RNAs and their implications in cancer epigenetics.
Beckedorff FC, Amaral MS, Deocesano-Pereira C, Verjovski-Almeida S. Beckedorff FC, et al. Biosci Rep. 2013 Aug 30;33(4):e00061. doi: 10.1042/BSR20130054. Biosci Rep. 2013. PMID: 23875687 Free PMC article. Review.
References
References and Notes
- Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10:155–159. - PubMed
- Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–641. - PubMed
- Sanchez-Elsner T, Gou D, Kremmer E, Sauer F. Noncoding RNAs of trithorax response elements recruit Drosophila Ash1 to Ultrabithorax. Science. 2006;311:1118–1123. - PubMed
References for Online-only Methods
- Bernstein BE, et al. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell. 2005;120:169–181. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials