Assembling a protein-protein interaction map of the SSU processome from existing datasets - PubMed (original) (raw)
Assembling a protein-protein interaction map of the SSU processome from existing datasets
Young H Lim et al. PLoS One. 2011.
Abstract
Background: The small subunit (SSU) processome is a large ribonucleoprotein complex involved in small ribosomal subunit assembly. It consists of the U3 snoRNA and ∼72 proteins. While most of its components have been identified, the protein-protein interactions (PPIs) among them remain largely unknown, and thus the assembly, architecture and function of the SSU processome remains unclear.
Methodology: We queried PPI databases for SSU processome proteins to quantify the degree to which the three genome-wide high-throughput yeast two-hybrid (HT-Y2H) studies, the genome-wide protein fragment complementation assay (PCA) and the literature-curated (LC) datasets cover the SSU processome interactome.
Conclusions: We find that coverage of the SSU processome PPI network is remarkably sparse. Two of the three HT-Y2H studies each account for four and six PPIs between only six of the 72 proteins, while the third study accounts for as little as one PPI and two proteins. The PCA dataset has the highest coverage among the genome-wide studies with 27 PPIs between 25 proteins. The LC dataset was the most extensive, accounting for 34 proteins and 38 PPIs, many of which were validated by independent methods, thereby further increasing their reliability. When the collected data were merged, we found that at least 70% of the predicted PPIs have yet to be determined and 26 proteins (36%) have no known partners. Since the SSU processome is conserved in all Eukaryotes, we also queried HT-Y2H datasets from six additional model organisms, but only four orthologues and three previously known interologous interactions were found. This provides a starting point for further work on SSU processome assembly, and spotlights the need for a more complete genome-wide Y2H analysis.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
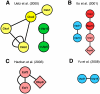
Figure 1. Interaction maps of the SSU processome proteins from existing HT-Y2H datasets.
Proteins are colored as described in the Materials and Methods; green nodes refer to proteins of the t-Utp/UtpA subcomplex, blue for UtpB, yellow for UtpC, gray for the U3 snoRNP proteins, brown for Bms1/Rcl1 and red for the Mpp10 subcomplex. Pink nodes refer to proteins that have yet to be assigned to a subcomplex. RNA helicases are depicted as diamonds. Multiple edges, or interactions, linking the proteins represent interactions identified in different studies or reciprocally identified as both bait and prey. Self-interactions are shown as looped edges. A) Results from the Uetz et al. dataset . B) Results from Ito et al. dataset . C) Results from the Hazbun et al. dataset . D) Results from the Yu et al. dataset .
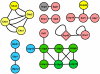
Figure 2. Interaction map of the SSU processome proteins from the PCA dataset.
Nodes are colored as in Fig. 1.
Figure 3. Interaction map of the SSU processome proteins from the LC dataset.
Nodes are depicted as in Fig. 1.
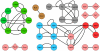
Figure 4. The current, merged SSU processome interactome map from the three HT-Y2H, PCA, LC and interologue datasets.
Interologues identified in Drosophila (D) and C. elegans (C) are also shown, with red and blue edges, respectively. The PPI redundancy (same interactions identified by different studies, methods or reciprocally) was removed from the figure to highlight the interacting partners. Nodes are depicted as in Fig. 1. Standalone nodes depict proteins without interaction data from any of the compiled datasets.
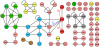
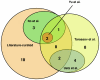
Figure 5. Comparison of the overlap between the HT-Y2H, PCA and LC datasets for the PPIs of the SSU processome.
Numbers within the Venn diagram refer to the number of SSU processome proteins present and overlapping in the HT-Y2H, PCA and LC datasets.
Similar articles
- The small subunit processome in ribosome biogenesis—progress and prospects.
Phipps KR, Charette J, Baserga SJ. Phipps KR, et al. Wiley Interdiscip Rev RNA. 2011 Jan-Feb;2(1):1-21. doi: 10.1002/wrna.57. Wiley Interdiscip Rev RNA. 2011. PMID: 21318072 Free PMC article. Review. - The SSU processome interactome in Saccharomyces cerevisiae reveals novel protein subcomplexes.
Vincent NG, Charette JM, Baserga SJ. Vincent NG, et al. RNA. 2018 Jan;24(1):77-89. doi: 10.1261/rna.062927.117. Epub 2017 Oct 20. RNA. 2018. PMID: 29054886 Free PMC article. - Bud23 promotes the final disassembly of the small subunit Processome in Saccharomyces cerevisiae.
Black JJ, Sardana R, Elmir EW, Johnson AW. Black JJ, et al. PLoS Genet. 2020 Dec 11;16(12):e1009215. doi: 10.1371/journal.pgen.1009215. eCollection 2020 Dec. PLoS Genet. 2020. PMID: 33306676 Free PMC article. - The DEAD-box RNA helicase-like Utp25 is an SSU processome component.
Charette JM, Baserga SJ. Charette JM, et al. RNA. 2010 Nov;16(11):2156-69. doi: 10.1261/rna.2359810. Epub 2010 Sep 30. RNA. 2010. PMID: 20884785 Free PMC article. - Human diseases of the SSU processome.
Sondalle SB, Baserga SJ. Sondalle SB, et al. Biochim Biophys Acta. 2014 Jun;1842(6):758-64. doi: 10.1016/j.bbadis.2013.11.004. Epub 2013 Nov 12. Biochim Biophys Acta. 2014. PMID: 24240090 Free PMC article. Review.
Cited by
- The small subunit processome in ribosome biogenesis—progress and prospects.
Phipps KR, Charette J, Baserga SJ. Phipps KR, et al. Wiley Interdiscip Rev RNA. 2011 Jan-Feb;2(1):1-21. doi: 10.1002/wrna.57. Wiley Interdiscip Rev RNA. 2011. PMID: 21318072 Free PMC article. Review. - Diversity in genetic in vivo methods for protein-protein interaction studies: from the yeast two-hybrid system to the mammalian split-luciferase system.
Stynen B, Tournu H, Tavernier J, Van Dijck P. Stynen B, et al. Microbiol Mol Biol Rev. 2012 Jun;76(2):331-82. doi: 10.1128/MMBR.05021-11. Microbiol Mol Biol Rev. 2012. PMID: 22688816 Free PMC article. Review. - hUTP24 is essential for processing of the human rRNA precursor at site A1, but not at site A0.
Tomecki R, Labno A, Drazkowska K, Cysewski D, Dziembowski A. Tomecki R, et al. RNA Biol. 2015;12(9):1010-29. doi: 10.1080/15476286.2015.1073437. RNA Biol. 2015. PMID: 26237581 Free PMC article. - Assembly and structure of the SSU processome-a nucleolar precursor of the small ribosomal subunit.
Barandun J, Hunziker M, Klinge S. Barandun J, et al. Curr Opin Struct Biol. 2018 Apr;49:85-93. doi: 10.1016/j.sbi.2018.01.008. Epub 2018 Feb 4. Curr Opin Struct Biol. 2018. PMID: 29414516 Free PMC article. Review. - A framework for application of metabolic modeling in yeast to predict the effects of nsSNV in human orthologs.
Dingerdissen H, Weaver DS, Karp PD, Pan Y, Simonyan V, Mazumder R. Dingerdissen H, et al. Biol Direct. 2014 Jun 3;9:9. doi: 10.1186/1745-6150-9-9. Biol Direct. 2014. PMID: 24894379 Free PMC article.
References
- Uetz P, Giot L, Cagney G, Mansfield TA, Judson RS, et al. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature. 2000;403:623–627. - PubMed
- Tarassov K, Messier V, Landry CR, Radinovic S, Serna Molina MM, et al. An in vivo map of the yeast protein interactome. Science. 2008;320:1465–1470. - PubMed
- Hazbun TR, Malmstrom L, Anderson S, Graczyk BJ, Fox B, et al. Assigning function to yeast proteins by integration of technologies. Mol Cell. 2003;12:1353–1365. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases