In vivo systems analysis identifies spatial and temporal aspects of the modulation of TNF-α-induced apoptosis and proliferation by MAPKs - PubMed (original) (raw)
In vivo systems analysis identifies spatial and temporal aspects of the modulation of TNF-α-induced apoptosis and proliferation by MAPKs
Ken S Lau et al. Sci Signal. 2011.
Abstract
Cellular responses to external stimuli depend on dynamic features of multipathway network signaling; thus, cell behavior is influenced in a complex manner by the environment and by intrinsic properties. Methods of multivariate systems analysis have provided an understanding of these convoluted effects, but only for relatively simplified examples in vitro. To determine whether such approaches could be successfully used in vivo, we analyzed the signaling network that determines the response of intestinal epithelial cells to tumor necrosis factor-α (TNF-α). We built data-driven, partial least-squares discriminant analysis (PLSDA) models based on signaling, apoptotic, and proliferative responses in the mouse small intestinal epithelium after systemic exposure to TNF-α. The extracellular signal-regulated kinase (ERK) signaling axis was a critical modulator of the temporal variation in apoptosis at different doses of TNF-α and of the spatial variation in proliferation in distinct intestinal regions. Inhibition of MEK, a mitogen-activated protein kinase kinase upstream of ERK, altered the signaling network and changed the temporal and spatial phenotypes consistent with model predictions. Our results demonstrate the dynamic, adaptive nature of in vivo signaling networks and identify natural, tissue-level variation in responses that can be deconvoluted only with quantitative, multivariate computational modeling. This study lays a foundation for the use of systems-based approaches to understand how dysregulation of the cellular network state underlies complex diseases.
Figures
Fig. 1
Regional dependence of the apoptotic and proliferative responses to TNF-α in the mouse small intestinal epithelium. (A) Morphological heterogeneity across the regions of the small intestine, as represented by H&E staining. The apoptotic response to TNF-α, as assessed by immunohistochemical analysis of cleaved caspase 3 (αCC3) at 4 hours after treatment with TNF-α (5 µg), and the proliferative response, as assessed by immunohistochemical analysis of phosphorylated histone H3 (αPH3) 30 min after treatment with TNF-α (5 µg), of the intestinal epithelium varies by region. (B) Schematic representation of the experimental design. After injection of mice with TNF-α (5 or 10 µg in PBS), protein lysates and tissue samples were obtained from the duodenum and ileum. In addition to 5 control animals that were mock-treated with PBS, 5 mice were analyzed for each dose of TNF-α at each time point (from 30 min to 8 hours). The Bio-Plex platform was used to generate phospho-protein signaling data, whereas quantitative Western blotting analysis was used to quantify apoptosis and total protein abundance. Immunohistochemical analysis was used to determine the localization of signals and to quantify the number of proliferating cells. (C) Time-course of apoptotic responses in the duodenum and ileum, as determined by quantitative Western blotting for CC3. Error bars represent the standard error of the mean (SEM) from five independent experiments. (D) Time-course of proliferative responses in the duodenum and ileum, as determined by enumerating the number of PH3-positive cells per crypt. Error bars represent the SEM from the numbers of crypts counted.
Fig. 2
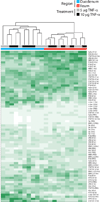
Time- and region-dependent signaling datasets in response to TNF-α. Hierarchical clustering of phosphorylated proteins measured by Bio-Plex. The intensity of the green color represents the average of technical duplicates of the median fluorescent intensity (normalized to the highest value of each signal) resulting from each assay. Regional and treatment groupings are signified by color at the top of the heat map. Data are compiled from five independent experiments. Phosphorylated protein signals corresponding to each row are listed on the right side of the heat map.
Fig. 3
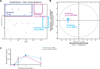
Statistical modeling of the in vivo signaling network describes the temporal and spatial variation in TNF-α-dependent phenotypes. (A) A PLSDA model constructed from the signaling dataset correlated with apoptosis and proliferation. Scores generated by the model. The dotted gray line represents the 95% confidence limit of the distribution of the scores. Three diverse phenotypic classes are delineated: late apoptosis and arrest (duodenum + 5 µg TNF-α, in cyan), early apoptosis and arrest (duodenum + 10 µg TNF-α, in blue), and no apoptosis or proliferation (ileum, in red). The percentages on the x-and y-axes represent the percentage of variance in the data set captured in a particular LV. (B) Loadings generated by the PLSDA model. Signals that contribute most to each phenotypic classification are labeled accordingly with the corresponding color scheme. (C) Representative signaling time courses for signals that distinguish between the classes, pc-JUN and pERK, from Bio-Plex analysis. Error bars represent the SEM from five independent experiments.
Fig. 4
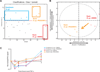
Model prediction and experimental test of the effect of MEK inhibition on TNF-α-induced apoptosis in the duodenum. (A) PLSDA model–based prediction of the apoptotic phenotype in the duodenum using phosphorylated protein abundance data from samples derived from animals treated with of TNF-α (5 µg) after pre-treatment with PD325901 (purple) or vehicle (10% DMSO in PBS, cyan), relative to samples from animals treated with 10 µg of TNF-α (blue). The y-axis shows the numerical result calculated with the PLSDA function of classification into the “early apoptosis” (class 2) phenotypic class. The red broken line is the threshold defining classification. (B) Scores plotted onto the LV1 and LV2 space of the PLSDA model showing the translocation of duodenal samples treated with TNF-α (5 µg) from the lower-left to the upper-left quadrant after MEK inhibition. (C) Experimental time-course of apoptosis induced in the duodenum by TNF-α (5 µg) after pretreatment with PD325901 (solid purple line) or vehicle (dotted cyan line). Error bars represent the SEM from three independent experiments.
Fig. 5
Model prediction and experimental test of the effect of MEK inhibition on TNF-α-induced proliferation in the ileum. (A) PLSDA model–based prediction of the classification into the “arrest” (class 1) phenotypic class using signaling data of ileal samples from mice treated with TNF-α (5 µg) after pre-treatment with PD325901 (orange) or vehicle (10% DMSO in PBS, red), relative to duodenal samples from mice treated with TNF-α (5 µg, cyan). (B) Scores plotted onto the LV1 and LV2 space of the PLSDA model showing the partial translocation of ileal samples treated with TNF-α-(5 µg) from the right half to the left half after inhibition of MEK. (C) Experimental time-course of proliferation induced by TNF-α (5 µg) in the duodenum and ileum after pretreatment with PD325901 (solid lines) or vehicle (dotted lines). Error bars represent the SEM from three independent experiments.
Fig. 6
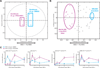
Inhibition of MEK initiates a global change in the TNF-α signaling network. (A) Scores of a 1D PLSDA model that separate TNF-α treatment (5 µg) in the duodenum with MEK inhibition (purple) from that with DMSO (cyan). The model is plotted in two dimensions for visual purposes. (B) Loadings of the model in (A). Signals that contribute most to each classification are labeled accordingly in the corresponding color scheme. (C) Abundances of specific phosphorylated proteins that most strongly distinguish the TNF-α-dependent phenotypes between the duodenal samples that were treated with PD325901 (solid purple line) from those that were treated with vehicle (dotted cyan line). Error bars represent the SEM from three independent experiments.
Fig. 7
The effect of local changes in the signaling network by inhibiting MEK. (A) PLSDA scores plot of an artificial duodenal dataset generated by setting the level of pERK to 0 in individual samples while leaving all other values the same (denoted as X on the plot). This dataset was compared to the original calibration dataset (cyan). Note that the artificial dataset does not shift. (B) PLSDA scores plot of an artificial dataset generated by setting the ratio of pMEK to pERK to 0, while leaving all other values the same, in ileal samples (X) of the calibration dataset, relative to the ileal samples of the original calibration dataset (red). Note that the artificial dataset shifts partially to the left.
Similar articles
- Kinase suppressor of Ras determines survival of intestinal epithelial cells exposed to tumor necrosis factor.
Yan F, John SK, Polk DB. Yan F, et al. Cancer Res. 2001 Dec 15;61(24):8668-75. Cancer Res. 2001. PMID: 11751383 - Herbal formula HMC05 prevents human aortic smooth muscle cell migration and proliferation by inhibiting the ERK1/2 MAPK signaling cascade.
Kang YH, Yang IJ, Shin HM. Kang YH, et al. J Nat Med. 2012 Jan;66(1):177-84. doi: 10.1007/s11418-011-0573-3. Epub 2011 Aug 11. J Nat Med. 2012. PMID: 21833774 - Silencing of human phosphatidylethanolamine-binding protein 4 sensitizes breast cancer cells to tumor necrosis factor-alpha-induced apoptosis and cell growth arrest.
Wang X, Li N, Li H, Liu B, Qiu J, Chen T, Cao X. Wang X, et al. Clin Cancer Res. 2005 Oct 15;11(20):7545-53. doi: 10.1158/1078-0432.CCR-05-0879. Clin Cancer Res. 2005. PMID: 16243830
Cited by
- Structural properties of the MAPK pathway topologies in PC12 cells.
Franco E, Blanchini F. Franco E, et al. J Math Biol. 2013 Dec;67(6-7):1633-68. doi: 10.1007/s00285-012-0606-x. Epub 2012 Oct 25. J Math Biol. 2013. PMID: 23096491 - TNF-insulin crosstalk at the transcription factor GATA6 is revealed by a model that links signaling and transcriptomic data tensors.
Chitforoushzadeh Z, Ye Z, Sheng Z, LaRue S, Fry RC, Lauffenburger DA, Janes KA. Chitforoushzadeh Z, et al. Sci Signal. 2016 Jun 7;9(431):ra59. doi: 10.1126/scisignal.aad3373. Sci Signal. 2016. PMID: 27273097 Free PMC article. - Functional and structural modifications of influenza antibodies during pregnancy.
Jennewein MF, Kosikova M, Noelette FJ, Radvak P, Boudreau CM, Campbell JD, Chen WH, Xie H, Alter G, Pasetti MF. Jennewein MF, et al. iScience. 2022 Mar 16;25(4):104088. doi: 10.1016/j.isci.2022.104088. eCollection 2022 Apr 15. iScience. 2022. PMID: 35402869 Free PMC article. - The extraction of simple relationships in growth factor-specific multiple-input and multiple-output systems in cell-fate decisions by backward elimination PLS regression.
Akimoto Y, Yugi K, Uda S, Kudo T, Komori Y, Kubota H, Kuroda S. Akimoto Y, et al. PLoS One. 2013 Sep 9;8(9):e72780. doi: 10.1371/journal.pone.0072780. eCollection 2013. PLoS One. 2013. PMID: 24039801 Free PMC article. - The colonic epithelium plays an active role in promoting colitis by shaping the tissue cytokine profile.
Lyons J, Ghazi PC, Starchenko A, Tovaglieri A, Baldwin KR, Poulin EJ, Gierut JJ, Genetti C, Yajnik V, Breault DT, Lauffenburger DA, Haigis KM. Lyons J, et al. PLoS Biol. 2018 Mar 29;16(3):e2002417. doi: 10.1371/journal.pbio.2002417. eCollection 2018 Mar. PLoS Biol. 2018. PMID: 29596476 Free PMC article.
References
- Bradley JR. TNF-mediated inflammatory disease. J. Pathol. 2008;214:149–160. - PubMed
- Rutgeerts P, Sandborn WJ, Feagan BG, Reinisch W, Olson A, Johanns J, Travers S, Rachmilewitz D, Hanauer SB, Lichtenstein GR, de Villiers WJ, Present D, Sands BE, Colombel JF. Infliximab for induction and maintenance therapy for ulcerative colitis. New Engl. J. Med. 2005;353:2462–2476. - PubMed
- Kontoyiannis D, Pasparakis M, Pizarro TT, Cominelli F, Kollias G. Impaired on/off regulation of TNF biosynthesis in mice lacking TNF AU-rich elements: implications for joint and gut-associated immunopathologies. Immunity. 1999;10:387–398. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- U54 CA112967/CA/NCI NIH HHS/United States
- U54-CA112967/CA/NCI NIH HHS/United States
- R01-GM088827/GM/NIGMS NIH HHS/United States
- R01 GM088827/GM/NIGMS NIH HHS/United States
- P30 DK043351/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous