An RNA interference-based screen of transcription factor genes identifies pathways necessary for sensory regeneration in the avian inner ear - PubMed (original) (raw)
An RNA interference-based screen of transcription factor genes identifies pathways necessary for sensory regeneration in the avian inner ear
David M Alvarado et al. J Neurosci. 2011.
Abstract
Sensory hair cells of the inner ear are the mechanoelectric transducers of sound and head motion. In mammals, damage to sensory hair cells leads to hearing or balance deficits. Nonmammalian vertebrates such as birds can regenerate hair cells after injury. In a previous study, we characterized transcription factor gene expression during chicken hair cell regeneration. In those studies, a laser microbeam or ototoxic antibiotics were used to damage the sensory epithelia (SE). The current study focused on 27 genes that were upregulated in regenerating SEs compared to untreated SEs in the previous study. Those genes were knocked down by siRNA to determine their requirement for supporting cell proliferation and to measure resulting changes in the larger network of gene expression. We identified 11 genes necessary for proliferation and also identified novel interactive relationships between many of them. Defined components of the WNT, PAX, and AP1 pathways were shown to be required for supporting cell proliferation. These pathways intersect on WNT4, which is also necessary for proliferation. Among the required genes, the CCAAT enhancer binding protein, CEBPG, acts downstream of Jun Kinase and JUND in the AP1 pathway. The WNT coreceptor LRP5 acts downstream of CEBPG, as does the transcription factor BTAF1. Both of these genes are also necessary for supporting cell proliferation. This is the first large-scale screen of its type and suggests an important intersection between the AP1 pathway, the PAX pathway, and WNT signaling in the regulation of supporting cell proliferation during inner ear hair cell regeneration.
Figures
Figure 1.
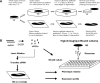
Experimental design. Flow diagram of experimental design scheme for time course profiling in the utricle and cochlea SE and RNAi profiling. A, Time course of laser and neomycin recovery. B, TFs revealed in the time course of recovery were targeted by siRNA to assess a proliferation phenotype and expression profiled to evaluate knockdown of the target gene and potential epistatic relationships between TFs.
Figure 2.
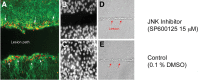
JNK signaling during SE regeneration. JNK signaling is evident at the leading edge of the lesion path in the SEs and necessary for proliferative regeneration. SE cultured on a glass coverslip was lesioned by microbeam laser ablation. A, Phosphorylated c-JUN was detected by a phosphorylation-specific antibody to the protein (red dots; white arrows). B, C, Following laser ablation, the cultured SE was treated with JNK inhibitor (SP600125, 15 μ
m
) (B) or 0.1% DMSO (control) (C) and allowed to recover for 24 h; nuclei are shown by DAPI staining. D, E, The laser lesion path is visible by etching of the coverslip through the phase contrast (D and E, red arrows). Only the JNK inhibitor exhibited a failure to close the wound.
Figure 3.
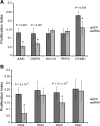
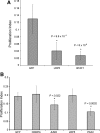
Effects of siRNA treatments on SC proliferation. Proliferation phenotypes were quantified for each siRNA knockdown compared to a GFP control by calculating a proliferation index. BrdU-labeled proliferating cells were compared to the total number of DAPI-stained cells to calculate a percentage proliferation for genes differentially expressed during SE regeneration (A) and PAX genes that were upregulated during SE regeneration (B).
Figure 4.
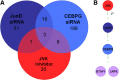
Analysis of overlapping expression profiles and novel epistatic relationships between genes that are necessary for SC proliferation. siRNA and inhibitor treatments were expression profiled to identify downstream effectors of SC proliferation. A, Numbers indicate genes differentially expressed in three treatments that each individually inhibit SC proliferation. Three genes are commonly downregulated, one of which is CEBPG. B, Novel epistatic relationships can be inferred from TF expression profiling siRNA and inhibitor treatments. CEBPG can be placed downstream of JNK and JUND and the other commonly downregulated genes, BTAF1 and LRP5.
Figure 5.
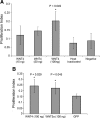
Analysis of siRNA treatments in chick SC and RPE proliferation. Percentage proliferation was quantified for siRNA treatments in chick SC for genes commonly downregulated in treatments that inhibit SC proliferation (downstream of the AP-1 pathway and CEBPG) (A) and chick RPE for genes that inhibited chick SC proliferation (B).
Figure 6.
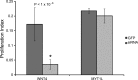
WNT4 and MYT1L siRNA phenotypes. WNT4 siRNA knockdowns inhibited SC proliferation compared to a GFP control, while MYT1L siRNA did not have a significant effect on proliferation.
Figure 7.
Exogenous WNT4 expression phenotypes. A, Percentage proliferation was quantified for treatment with 15, 50, or 100 ng/well of exogenous WNT4 protein. Cells treated with 100 ng/well of exogenous WNT4 protein hyperproliferated compared to heat-inactivated WNT4 and cells cultured in unsupplemented media. B, Treatment with exogenous WNT5a also caused hyperproliferation; however, CEBPG siRNA in combination with exogenous WNT4 treatment inhibited proliferation similarly to CEBPG siRNA treatment alone.
Similar articles
- The transcriptome of utricle hair cell regeneration in the avian inner ear.
Ku YC, Renaud NA, Veile RA, Helms C, Voelker CC, Warchol ME, Lovett M. Ku YC, et al. J Neurosci. 2014 Mar 5;34(10):3523-35. doi: 10.1523/JNEUROSCI.2606-13.2014. J Neurosci. 2014. PMID: 24599453 Free PMC article. - Musashi-1 is the candidate of the regulator of hair cell progenitors during inner ear regeneration.
Wakasaki T, Niiro H, Jabbarzadeh-Tabrizi S, Ohashi M, Kimitsuki T, Nakagawa T, Komune S, Akashi K. Wakasaki T, et al. BMC Neurosci. 2017 Aug 16;18(1):64. doi: 10.1186/s12868-017-0382-z. BMC Neurosci. 2017. PMID: 28814279 Free PMC article. - YAP Mediates Hair Cell Regeneration in Balance Organs of Chickens, But LATS Kinases Suppress Its Activity in Mice.
Rudolf MA, Andreeva A, Kozlowski MM, Kim CE, Moskowitz BA, Anaya-Rocha A, Kelley MW, Corwin JT. Rudolf MA, et al. J Neurosci. 2020 May 13;40(20):3915-3932. doi: 10.1523/JNEUROSCI.0306-20.2020. Epub 2020 Apr 27. J Neurosci. 2020. PMID: 32341094 Free PMC article. - Sensory regeneration in the vertebrate inner ear: differences at the levels of cells and species.
Warchol ME. Warchol ME. Hear Res. 2011 Mar;273(1-2):72-9. doi: 10.1016/j.heares.2010.05.004. Epub 2010 May 19. Hear Res. 2011. PMID: 20488231 Review. - Atoh1 and other related key regulators in the development of auditory sensory epithelium in the mammalian inner ear: function and interplay.
Zhong C, Fu Y, Pan W, Yu J, Wang J. Zhong C, et al. Dev Biol. 2019 Feb 15;446(2):133-141. doi: 10.1016/j.ydbio.2018.12.025. Epub 2018 Dec 31. Dev Biol. 2019. PMID: 30605626 Review.
Cited by
- Making sense of Wnt signaling-linking hair cell regeneration to development.
Jansson L, Kim GS, Cheng AG. Jansson L, et al. Front Cell Neurosci. 2015 Mar 11;9:66. doi: 10.3389/fncel.2015.00066. eCollection 2015. Front Cell Neurosci. 2015. PMID: 25814927 Free PMC article. Review. - Responses to cell loss become restricted as the supporting cells in mammalian vestibular organs grow thick junctional actin bands that develop high stability.
Burns JC, Corwin JT. Burns JC, et al. J Neurosci. 2014 Jan 29;34(5):1998-2011. doi: 10.1523/JNEUROSCI.4355-13.2014. J Neurosci. 2014. PMID: 24478379 Free PMC article. - Regeneration in the Auditory Organ in Cuban and African Dwarf Crocodiles (Crocodylus rhombifer and Osteolaemus tetraspis) Can We Learn From the Crocodile How to Restore Our Hearing?
Li H, Staxäng K, Hodik M, Melkersson KG, Rask-Andersen M, Rask-Andersen H. Li H, et al. Front Cell Dev Biol. 2022 Jul 4;10:934571. doi: 10.3389/fcell.2022.934571. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 35859896 Free PMC article. - In vitro and in vivo models: What have we learnt about inner ear regeneration and treatment for hearing loss?
Lee MP, Waldhaus J. Lee MP, et al. Mol Cell Neurosci. 2022 May;120:103736. doi: 10.1016/j.mcn.2022.103736. Epub 2022 May 14. Mol Cell Neurosci. 2022. PMID: 35577314 Free PMC article. Review. - The transcriptome of utricle hair cell regeneration in the avian inner ear.
Ku YC, Renaud NA, Veile RA, Helms C, Voelker CC, Warchol ME, Lovett M. Ku YC, et al. J Neurosci. 2014 Mar 5;34(10):3523-35. doi: 10.1523/JNEUROSCI.2606-13.2014. J Neurosci. 2014. PMID: 24599453 Free PMC article.
References
- Adam J, Myat A, Le Roux I, Eddison M, Henrique D, Ish-Horowicz D, Lewis J. Cell fate choices and the expression of Notch, Delta and Serrate homologues in the chick inner ear: parallels with Drosophila sense-organ development. Development. 1998;125:4645–4654. - PubMed
- Brivanlou AH, Darnell JE., Jr Signal transduction and the control of gene expression. Science. 2002;295:813–818. - PubMed
- Chang J, Sonoyama W, Wang Z, Jin Q, Zhang C, Krebsbach PH, Giannobile W, Shi S, Wang CY. Noncanonical Wnt-4 signaling enhances bone regeneration of mesenchymal stem cells in craniofacial defects through activation of p38 MAPK. J Biol Chem. 2007;282:30938–30948. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P30 DC004665/DC/NIDCD NIH HHS/United States
- R01 DC006283/DC/NIDCD NIH HHS/United States
- R01 DC005632/DC/NIDCD NIH HHS/United States
- R01 DC006283-07/DC/NIDCD NIH HHS/United States
- P30 DC004665-08/DC/NIDCD NIH HHS/United States
- R01DC006283/DC/NIDCD NIH HHS/United States
- R01 DC005632-04/DC/NIDCD NIH HHS/United States
- RC1 DC010677-01/DC/NIDCD NIH HHS/United States
- P30DC04665/DC/NIDCD NIH HHS/United States
- R01DC005632/DC/NIDCD NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous