A cis-regulatory map of the Drosophila genome - PubMed (original) (raw)
. 2011 Mar 24;471(7339):527-31.
doi: 10.1038/nature09990.
Christopher D Brown, Lijia Ma, Christopher Aaron Bristow, Steven W Miller, Ulrich Wagner, Pouya Kheradpour, Matthew L Eaton, Paul Loriaux, Rachel Sealfon, Zirong Li, Haruhiko Ishii, Rebecca F Spokony, Jia Chen, Lindsay Hwang, Chao Cheng, Richard P Auburn, Melissa B Davis, Marc Domanus, Parantu K Shah, Carolyn A Morrison, Jennifer Zieba, Sarah Suchy, Lionel Senderowicz, Alec Victorsen, Nicholas A Bild, A Jason Grundstad, David Hanley, David M MacAlpine, Mattias Mannervik, Koen Venken, Hugo Bellen, Robert White, Mark Gerstein, Steven Russell, Robert L Grossman, Bing Ren, James W Posakony, Manolis Kellis, Kevin P White
Affiliations
- PMID: 21430782
- PMCID: PMC3179250
- DOI: 10.1038/nature09990
A cis-regulatory map of the Drosophila genome
Nicolas Nègre et al. Nature. 2011.
Abstract
Systematic annotation of gene regulatory elements is a major challenge in genome science. Direct mapping of chromatin modification marks and transcriptional factor binding sites genome-wide has successfully identified specific subtypes of regulatory elements. In Drosophila several pioneering studies have provided genome-wide identification of Polycomb response elements, chromatin states, transcription factor binding sites, RNA polymerase II regulation and insulator elements; however, comprehensive annotation of the regulatory genome remains a significant challenge. Here we describe results from the modENCODE cis-regulatory annotation project. We produced a map of the Drosophila melanogaster regulatory genome on the basis of more than 300 chromatin immunoprecipitation data sets for eight chromatin features, five histone deacetylases and thirty-eight site-specific transcription factors at different stages of development. Using these data we inferred more than 20,000 candidate regulatory elements and validated a subset of predictions for promoters, enhancers and insulators in vivo. We identified also nearly 2,000 genomic regions of dense transcription factor binding associated with chromatin activity and accessibility. We discovered hundreds of new transcription factor co-binding relationships and defined a transcription factor network with over 800 potential regulatory relationships.
Figures
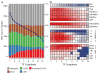
Figure 1. Chromatin dynamics across Drosophila development
(A) Enrichment of CBP and H3K4me1 (rows) within regions marked by other chromatin modifications, factors, or annotated enhancers (columns). Note that (i) CBP is enriched within all active marks (H3K4me3, H3K27Ac, H3K9Ac, H3K4me1 and PolII) at all stages of development and (ii) early embryo (0–16h) CBP and H3K4me1 marked regions are enriched within H3K27me3 domains and annotated enhancers (right panel). (B) Heatmap depicting fold enrichment of CBP bound regions (columns) at different developmental stages for each of the 22 clusters of TSS-distal regions (rows) grouped by their protein binding profiles. A subset of the clusters shows significant enrichment for CBP at different developmental stages. (C) Enrichment of enhancer categories (columns) for each of the 22 clusters of TSS-distal regions (rows). Many clusters enriched for CBP binding in early development are also strongly enriched for enhancers (rows with *). (D) Embryo-specific CBP binding predicts unannotated enhancers. RNA in situs with a Gal4 probe were used to stain transgenic embryos representing five different enhancer predictions (rows), at four to five different stages (columns). EO044 overlaps the known expression pattern for the neighboring gene, CG8745 (FlyExpress Database). (E) Enrichment of enhancer annotations (rows) within the binding sites of each transcription factor (columns). For panels C and G gray boxes indicate no overlap. For panels D and E all values greater or less than zero are significant, FDR < 0.01.
Figure 2. Transcription factor binding site complexity
(A) Number of TFBS (left y-axis, black circles) and distribution of genomic annotation classes (right y-axis, colors) as a function of TFBS complexity (x-axis). (B) TFBS enrichment (color scale, depleted in blue, enriched in red) of TFBS sorted by TF binding site complexity (x-axis) within annotated enhancers (CM: cardiac mesoderm, Ht: heart muscle, SM: somatic muscle; VM: visceral muscle.), HDAC binding sites, early embryo chromatin marks. At top is a heatmap depicting nucleosome density as a function of TFBS complexity.
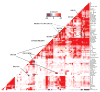
Figure 3. Transcription factor binding site overlap
Pairwise TFBS enrichment/depletion (colour coded by Z-score). TFBS datasets labeled at left. Selected interactions described in the text are highlighted.
Comment in
- Molecular biology: A fly in the face of genomics.
Furlong EE. Furlong EE. Nature. 2011 Mar 24;471(7339):458-9. doi: 10.1038/471458a. Nature. 2011. PMID: 21430772 No abstract available.
Similar articles
- Diverse patterns of genomic targeting by transcriptional regulators in Drosophila melanogaster.
Slattery M, Ma L, Spokony RF, Arthur RK, Kheradpour P, Kundaje A, Nègre N, Crofts A, Ptashkin R, Zieba J, Ostapenko A, Suchy S, Victorsen A, Jameel N, Grundstad AJ, Gao W, Moran JR, Rehm EJ, Grossman RL, Kellis M, White KP. Slattery M, et al. Genome Res. 2014 Jul;24(7):1224-35. doi: 10.1101/gr.168807.113. Genome Res. 2014. PMID: 24985916 Free PMC article. - Genome-Wide Mapping Targets of the Metazoan Chromatin Remodeling Factor NURF Reveals Nucleosome Remodeling at Enhancers, Core Promoters and Gene Insulators.
Kwon SY, Grisan V, Jang B, Herbert J, Badenhorst P. Kwon SY, et al. PLoS Genet. 2016 Apr 5;12(4):e1005969. doi: 10.1371/journal.pgen.1005969. eCollection 2016 Apr. PLoS Genet. 2016. PMID: 27046080 Free PMC article. - The Ubx Polycomb response element bypasses an unpaired Fab-8 insulator via cis transvection in Drosophila.
Lu D, Li Z, Li L, Yang L, Chen G, Yang D, Zhang Y, Singh V, Smith S, Xiao Y, Wang E, Ye Y, Zhang W, Zhou L, Rong Y, Zhou J. Lu D, et al. PLoS One. 2018 Jun 21;13(6):e0199353. doi: 10.1371/journal.pone.0199353. eCollection 2018. PLoS One. 2018. PMID: 29928011 Free PMC article. - Regulatory regions in DNA: promoters, enhancers, silencers, and insulators.
Riethoven JJ. Riethoven JJ. Methods Mol Biol. 2010;674:33-42. doi: 10.1007/978-1-60761-854-6_3. Methods Mol Biol. 2010. PMID: 20827584 Review. - Chromatin insulators.
Valenzuela L, Kamakaka RT. Valenzuela L, et al. Annu Rev Genet. 2006;40:107-38. doi: 10.1146/annurev.genet.39.073003.113546. Annu Rev Genet. 2006. PMID: 16953792 Review.
Cited by
- Identifying targets of the Sox domain protein Dichaete in the Drosophila CNS via targeted expression of dominant negative proteins.
Shen SP, Aleksic J, Russell S. Shen SP, et al. BMC Dev Biol. 2013 Jan 5;13:1. doi: 10.1186/1471-213X-13-1. BMC Dev Biol. 2013. PMID: 23289785 Free PMC article. - One Hand Clapping: detection of condition-specific transcription factor interactions from genome-wide gene activity data.
Dümcke S, Seizl M, Etzold S, Pirkl N, Martin DE, Cramer P, Tresch A. Dümcke S, et al. Nucleic Acids Res. 2012 Oct;40(18):8883-92. doi: 10.1093/nar/gks695. Epub 2012 Jul 25. Nucleic Acids Res. 2012. PMID: 22844089 Free PMC article. - Cis-regulatory chromatin loops arise before TADs and gene activation, and are independent of cell fate during early Drosophila development.
Espinola SM, Götz M, Bellec M, Messina O, Fiche JB, Houbron C, Dejean M, Reim I, Cardozo Gizzi AM, Lagha M, Nollmann M. Espinola SM, et al. Nat Genet. 2021 Apr;53(4):477-486. doi: 10.1038/s41588-021-00816-z. Epub 2021 Apr 1. Nat Genet. 2021. PMID: 33795867 - Systematic evaluation of factors influencing ChIP-seq fidelity.
Chen Y, Negre N, Li Q, Mieczkowska JO, Slattery M, Liu T, Zhang Y, Kim TK, He HH, Zieba J, Ruan Y, Bickel PJ, Myers RM, Wold BJ, White KP, Lieb JD, Liu XS. Chen Y, et al. Nat Methods. 2012 Jun;9(6):609-14. doi: 10.1038/nmeth.1985. Epub 2012 Apr 22. Nat Methods. 2012. PMID: 22522655 Free PMC article. - The insulator protein Suppressor of Hairy wing is required for proper ring canal development during oogenesis in Drosophila.
Hsu SJ, Plata MP, Ernest B, Asgarifar S, Labrador M. Hsu SJ, et al. Dev Biol. 2015 Jul 1;403(1):57-68. doi: 10.1016/j.ydbio.2015.03.024. Epub 2015 Apr 14. Dev Biol. 2015. PMID: 25882370 Free PMC article.
References
- Ren B, et al. Genome-wide location and function of DNA binding proteins. Science. 2000;290 (5500):2306–2309. - PubMed
- Johnson DS, Mortazavi A, Myers RM, Wold B. Genome-wide mapping of in vivo protein-DNA interactions. Science. 2007;316 (5830):1497–1502. - PubMed
- Schuettengruber B, Chourrout D, Vervoort M, Leblanc B, Cavalli G. Genome regulation by polycomb and trithorax proteins. Cell. 2007;128 (4):735–745. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- F32 GM074364-02/GM/NIGMS NIH HHS/United States
- P50 GM081892/GM/NIGMS NIH HHS/United States
- R01 HG004037/HG/NHGRI NIH HHS/United States
- R01 HG004037-04/HG/NHGRI NIH HHS/United States
- F32 GM074364-01/GM/NIGMS NIH HHS/United States
- RC2 HG005639-02/HG/NHGRI NIH HHS/United States
- U01 HG004279/HG/NHGRI NIH HHS/United States
- U01HG004264/HG/NHGRI NIH HHS/United States
- RC2 HG005639/HG/NHGRI NIH HHS/United States
- U01 HG004264/HG/NHGRI NIH HHS/United States
- F32 GM074364/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases