Genomic views of STAT function in CD4+ T helper cell differentiation - PubMed (original) (raw)
Review
Genomic views of STAT function in CD4+ T helper cell differentiation
John J O'Shea et al. Nat Rev Immunol. 2011 Apr.
Abstract
Signal transducer and activator of transcription (STAT) proteins are well known for their essential roles in transmitting cytokine-mediated signals and specifying T helper (T(H)) cell differentiation. Recent technological advances have revealed that STAT proteins have broad and complex roles in gene regulation and epigenetic control, including important roles as functional repressors. However, the challenge of how to link signal transduction, nucleosome biology and gene regulation remains. The relevance of tackling this problem is highlighted by genome-wide association studies that link cytokine signalling and STATs to various autoimmune or immune deficiency disorders. Defining exactly how extrinsic signals control the specification and plasticity of T(H) cells will provide important insights and perhaps therapeutic opportunities in these diseases.
Figures
Figure 1. Experimental flow of ChIP-seq analysis
A technique to combine chromatin immunoprecipitation with next generation sequencing to map DNA–protein interactions across the whole genome is shown. Chemically cross-linked DNA-protein complexes are immunoprecipitated and the protein-bound DNA fragments are isolated. The crosslinks are reversed, and the purified DNA was used to generate a library for sequencing. Automated reactions yield 36 nt long sequence reads of over 20 million per sample (Illumina GA platform). The sequence reads are aligned onto the reference genome and the distribution of protein-DNA interaction sites are visualized as “peaks” on the genome browser.
Figure 2. Distinctive epigenetic patterns are formed by STAT proteins in differentiated T helper effector cells
A key role of signal transducer and activator of transcription (STAT) proteins includes shaping epigenetic patterns on target gene loci to maintain cell lineage specificity. Five distinct epigenetic patterns were found to be STAT4 dependent in T helper 1 (TH1) cells that included both permissive chromatin signatures (high histone 3 lysine 4 trimethylation (H3K4me3) marks, high H3K36me3 marks or low H3K27me3 marks) and repressive chromatin signatures (high H3K27me3 or low H3K36me3). Permissive chromatin signatures are found on TH1 cell-expressed genes, whereas repressed chromatin signatures are found on TH2 cell-expressed genes in TH1 cells. The figure was reproduced from online version of Wei et al, Immunity 2010.
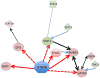
Figure 3. The STAT6 signalling network identified during the initial TH2 cell differentiation stage
Interleukin-4 (IL-4)- and signal transducer and activator of transcription 6 (STAT6)-regulated transcription factors form a core network of interacting nodes. Genes shown in red color boxes are upregulated and those in green color boxes are downregulated by STAT6 in transcriptomics studies. STAT6-mediated regulation of genes detected by ChIP-seq is marked with red arrows (solid line for direct regulation, dashed line for indirect regulation). Furthermore, known direct interactions between the putative downstream transcriptional regulators of STAT6 in humans were added to the figure. Blue lines correspond to protein-protein interactions, and black lines correspond to other types of interaction or regulation. The networks were generated through the use of Ingenuity Pathways Analysis (Ingenuity Systems,
) with some modifications based on published reports. The figure was modified from Elo et al. Immunity 2010 with the permission from Immunity.
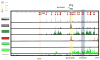
Figure 4. Markers of genomic organization to define activities of chromosome regions
Genomic organization encompassing the interferon-g (Ifng) locus in T helper (TH) cells. In TH1 cells, in which the Ifng locus is actively transcribed in a signal transducer and activator of transcription 4 (STAT4)-dependent manner, the promoter is marked by permissive histone 3 lysine 4 trimethylation (H3K4me3) and STAT4 binding, and the gene body is marked by permissive H3K4me3 and H3K36me3 marks. One of the distal enhancer elements (shaded in gray) is marked by H3K4me1 and STAT4 binding in TH1 cells and by repressive H3K27me3 in TH2 cells. Further 5′ upstream of the Ifng locus, an insulator site marked by CTCF binding is located and all permissive histone marks and DNase hypersensitivity sites are restricted beyond that point. Components of the JAK–STAT signaling pathway have been identified as causal genes for autoimmune diseases and have also been implicated in genetic linkage studies as having statistically significant differences between patients and controls.
Similar articles
- Helper T-cell differentiation and plasticity: insights from epigenetics.
Hirahara K, Vahedi G, Ghoreschi K, Yang XP, Nakayamada S, Kanno Y, O'Shea JJ, Laurence A. Hirahara K, et al. Immunology. 2011 Nov;134(3):235-45. doi: 10.1111/j.1365-2567.2011.03483.x. Immunology. 2011. PMID: 21977994 Free PMC article. Review. - JAK/STAT disruption induces immuno-deficiency: Rationale for the development of JAK inhibitors as immunosuppressive drugs.
Cornez I, Yajnanarayana SP, Wolf AM, Wolf D. Cornez I, et al. Mol Cell Endocrinol. 2017 Aug 15;451:88-96. doi: 10.1016/j.mce.2017.01.035. Epub 2017 Jan 25. Mol Cell Endocrinol. 2017. PMID: 28131742 Review. - Expansion of T follicular helper-T helper 1 like cells through epigenetic regulation by signal transducer and activator of transcription factors.
Ma X, Nakayamada S, Kubo S, Sakata K, Yamagata K, Miyazaki Y, Yoshikawa M, Kitanaga Y, Zhang M, Tanaka Y. Ma X, et al. Ann Rheum Dis. 2018 Sep;77(9):1354-1361. doi: 10.1136/annrheumdis-2017-212652. Epub 2018 May 31. Ann Rheum Dis. 2018. PMID: 29853448 - [Effector function plasticity of T helper lymphocytes].
Frey O, Kamradt T. Frey O, et al. Z Rheumatol. 2009 Dec;68(10):834-5. doi: 10.1007/s00393-009-0559-7. Z Rheumatol. 2009. PMID: 19847446 Review. German. - The Current STATus of lymphocyte signaling: new roles for old players.
Adamson AS, Collins K, Laurence A, O'Shea JJ. Adamson AS, et al. Curr Opin Immunol. 2009 Apr;21(2):161-6. doi: 10.1016/j.coi.2009.03.013. Epub 2009 Apr 9. Curr Opin Immunol. 2009. PMID: 19362457 Free PMC article. Review.
Cited by
- Immune perturbations in human pancreas lymphatic tissues prior to and after type 1 diabetes onset.
Golden GJ, Wu VH, Hamilton JT, Amses KR, Shapiro MR, Japp AS, Liu C, Pampena MB, Kuri-Cervantes L, Knox JJ, Gardner JS; HPAP Consortium; Atkinson MA, Brusko TM, Prak ETL, Kaestner KH, Naji A, Betts MR. Golden GJ, et al. bioRxiv [Preprint]. 2024 Sep 16:2024.04.23.590798. doi: 10.1101/2024.04.23.590798. bioRxiv. 2024. PMID: 39345402 Free PMC article. Preprint. - CBP/P300 Inhibition Impairs CD4+ T Cell Activation: Implications for Autoimmune Disorders.
Picavet LW, Samat AAK, Calis J, Nijhuis L, Scholman R, Mokry M, Tough DF, Prinjha RK, Vastert SJ, van Loosdregt J. Picavet LW, et al. Biomedicines. 2024 Jun 18;12(6):1344. doi: 10.3390/biomedicines12061344. Biomedicines. 2024. PMID: 38927552 Free PMC article. - HIF2α-dependent Dock4/Rac1-signaling regulates formation of adherens junctions and cell polarity in normoxia.
Raykhel I, Ronkainen VP, Myllyharju J, Manninen A. Raykhel I, et al. Sci Rep. 2024 May 27;14(1):12153. doi: 10.1038/s41598-024-62955-7. Sci Rep. 2024. PMID: 38802496 Free PMC article. - Regulation of T helper cell differentiation by the interplay between histone modification and chromatin interaction.
Liu S, Cao Y, Cui K, Ren G, Zhao T, Wang X, Wei D, Chen Z, Gurram RK, Liu C, Wu C, Zhu J, Zhao K. Liu S, et al. Immunity. 2024 May 14;57(5):987-1004.e5. doi: 10.1016/j.immuni.2024.03.018. Epub 2024 Apr 12. Immunity. 2024. PMID: 38614090 - Deciphering the developmental trajectory of tissue-resident Foxp3+ regulatory T cells.
Alvarez F, Liu Z, Bay A, Piccirillo CA. Alvarez F, et al. Front Immunol. 2024 Mar 28;15:1331846. doi: 10.3389/fimmu.2024.1331846. eCollection 2024. Front Immunol. 2024. PMID: 38605970 Free PMC article. Review.
References
- Littman DR, Rudensky AY. Th17 and regulatory T cells in mediating and restraining inflammation. Cell. 2010;140:845–58. - PubMed
- Korn T, Bettelli E, Oukka M, Kuchroo VK. IL-17 and Th17 Cells. Annu Rev Immunol. 2009;27:485–517. - PubMed
- Veldhoen M, Hocking RJ, Atkins CJ, Locksley RM, Stockinger B. TGFbeta in the context of an inflammatory cytokine milieu supports de novo differentiation of IL-17-producing T cells. Immunity. 2006;24:179–89. - PubMed
- Veldhoen M, et al. Transforming growth factor-beta ‘reprograms’ the differentiation of T helper 2 cells and promotes an interleukin 9-producing subset. Nat Immunol. 2008;9:1341–6. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials