A molecular phylogeny of living primates - PubMed (original) (raw)
. 2011 Mar;7(3):e1001342.
doi: 10.1371/journal.pgen.1001342. Epub 2011 Mar 17.
Warren E Johnson, Christian Roos, Hector N Seuánez, Julie E Horvath, Miguel A M Moreira, Bailey Kessing, Joan Pontius, Melody Roelke, Yves Rumpler, Maria Paula C Schneider, Artur Silva, Stephen J O'Brien, Jill Pecon-Slattery
Affiliations
- PMID: 21436896
- PMCID: PMC3060065
- DOI: 10.1371/journal.pgen.1001342
A molecular phylogeny of living primates
Polina Perelman et al. PLoS Genet. 2011 Mar.
Abstract
Comparative genomic analyses of primates offer considerable potential to define and understand the processes that mold, shape, and transform the human genome. However, primate taxonomy is both complex and controversial, with marginal unifying consensus of the evolutionary hierarchy of extant primate species. Here we provide new genomic sequence (~8 Mb) from 186 primates representing 61 (~90%) of the described genera, and we include outgroup species from Dermoptera, Scandentia, and Lagomorpha. The resultant phylogeny is exceptionally robust and illuminates events in primate evolution from ancient to recent, clarifying numerous taxonomic controversies and providing new data on human evolution. Ongoing speciation, reticulate evolution, ancient relic lineages, unequal rates of evolution, and disparate distributions of insertions/deletions among the reconstructed primate lineages are uncovered. Our resolution of the primate phylogeny provides an essential evolutionary framework with far-reaching applications including: human selection and adaptation, global emergence of zoonotic diseases, mammalian comparative genomics, primate taxonomy, and conservation of endangered species.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
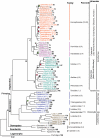
Figure 1. The molecular phylogeny of 61 Primate genera, two Dermoptera genera, and one Scandentia genus and rooted by Lagomorpha.
Shown is the maximum likelihood tree based on 34,927 bp sequenced from 54 genes amplified from selected single species representing each genus. All unmarked nodes have bootstrap support of 100%. Nodes with green circles have bootstrap proportions<70%, grey circles 71–80%, black circles 81–90% and red circles 91–99%. Boxes indicate genus of species with completed, nominated or draft whole genome sequence accomplished. Numbers in parenthesis next to each genus indicate number of species present in study followed by the total number described . Numbers in parentheses next to family names indicate number of genera included in study followed by total described . Numbers in bold refer to nodes on Figure 2, Figure S1, Table 1, Table 2, Table 3. Reference fossil dates used for calibration of tree in dating algorithms are represented by letters A-H on nodes (see Materials and Methods). Fossil dates are as follows and sources are listed in Materials and Methods: A) Galagidae-Lorisidae split 38–42 MYA, B) Simiiformes emerge 36–50 MYA, C) Catarrhini emerge 20–38 MYA, D) Platyrrhini emerge 20–27 MYA, E) Tribe Papionini emerge 6–8 MYA, F) Theropithecus emerge 3.5–4.5 MYA, G) Family Hominidae emerge 13–18 MYA, H) Homo-Pan split 6–7 MYA.
Figure 2. The molecular phylogeny of 186 primates and four species representing the two outgroup orders of Scandentia, Dermoptera, and rooted by Lagomorpha.
(See also Figure S1). Shown is the maximum likelihood tree derived from 34,927 bp of sequence from 54 genes. Node support is >90% for 166 nodes. Each node within the tree is numbered and listed in Table 1, Table 2, Table 3 to provide all node support values for ML, MP and Bayesian methods of analysis as well as estimated dates of divergence. Numbers in boxes represent estimate divergence times for major nodes as listed in Table 1, Table 2, Table 3. * denotes nodes whose divergence time is estimated to be less than 1 MYA.
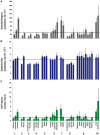
Figure 3. Patterns of nucleotide substitution and indel frequency in different categories of primate taxonomy.
- infraorders Simiiformes, Lemuriformes, and Lorisiformes (Chiromyiformes and Tarsiiformes excluded due to small numbers of species); 2) parvorders Catarrhini and Platyrrhini; 3) superfamilies Cercopithecoidea and Hominoidea; 4) catarrhine families Cercopithecidae, Hominidae and Hylobatidae, 5) platyrrhine families Pitheciidae Atelidae, and Cebidae; 6) Malagasy strepsirrhine families of Lemuridae, Indriidae, Lepilemuridae, and Cheirogaleidae; 7) strepsirrhine families of Lorisidae and Galagidae; 8) catarrhine subfamilies of Cercopithecinae, Colobinae, Homininae, and Ponginae; 9) platyrrhine subfamilies of Callitrichinae, Aotinae, Cebinae, Saimirinae, Alouattinae, Atelidae, Calicebinae and Pitheciinae; 10) strepsirrhine subfamilies of Lorisinae and Perodicticinae. (A) Mean nucleotide divergence and standard error computed from branch lengths per taxonomic level from Figure 2, Figure S1, Table 1, Table 2, Table 3, and Tables S6, S7, S8. (B) Mean rate of nucleotide substitution and standard error computed from BEAST analysis for each branch within taxonomic level from Table 1, Table 2, Table 3, and Tables S6, S7, S8. (C) Mean number of synapomorphic and autapomorphic indels per branch and standard error computed from Table 1, Table 2, Table 3, and Tables S6, S7, S8. Horizontal lines reflect global mean for primate phylogeny for each parameter.
Comment in
- Phylogenomics: improving our family tree.
Casci T. Casci T. Nat Rev Genet. 2011 May;12(5):298. doi: 10.1038/nrg2996. Epub 2011 Apr 6. Nat Rev Genet. 2011. PMID: 21468100 No abstract available.
Similar articles
- Recent advances in primate phylogenomics.
Pecon-Slattery J. Pecon-Slattery J. Annu Rev Anim Biosci. 2014 Feb;2:41-63. doi: 10.1146/annurev-animal-022513-114217. Annu Rev Anim Biosci. 2014. PMID: 25384134 Review. - Molecular and genomic data identify the closest living relative of primates.
Janecka JE, Miller W, Pringle TH, Wiens F, Zitzmann A, Helgen KM, Springer MS, Murphy WJ. Janecka JE, et al. Science. 2007 Nov 2;318(5851):792-4. doi: 10.1126/science.1147555. Science. 2007. PMID: 17975064 - Moving primate genomics beyond the chimpanzee genome.
Goodman M, Grossman LI, Wildman DE. Goodman M, et al. Trends Genet. 2005 Sep;21(9):511-7. doi: 10.1016/j.tig.2005.06.012. Trends Genet. 2005. PMID: 16009448 Review. - Comparative RNA sequencing reveals substantial genetic variation in endangered primates.
Perry GH, Melsted P, Marioni JC, Wang Y, Bainer R, Pickrell JK, Michelini K, Zehr S, Yoder AD, Stephens M, Pritchard JK, Gilad Y. Perry GH, et al. Genome Res. 2012 Apr;22(4):602-10. doi: 10.1101/gr.130468.111. Epub 2011 Dec 29. Genome Res. 2012. PMID: 22207615 Free PMC article. - The contribution of admixture to primate evolution.
Tung J, Barreiro LB. Tung J, et al. Curr Opin Genet Dev. 2017 Dec;47:61-68. doi: 10.1016/j.gde.2017.08.010. Epub 2017 Sep 15. Curr Opin Genet Dev. 2017. PMID: 28923540 Review.
Cited by
- Contagious yawning and scratching in captive lemurs.
Lemes WP, Amici F. Lemes WP, et al. Sci Rep. 2024 Nov 4;14(1):26672. doi: 10.1038/s41598-024-77805-9. Sci Rep. 2024. PMID: 39496688 Free PMC article. - The evolution of gestation length in eutherian mammals.
Danis T, Rokas A. Danis T, et al. Proc Biol Sci. 2024 Oct;291(2033):20241412. doi: 10.1098/rspb.2024.1412. Epub 2024 Oct 30. Proc Biol Sci. 2024. PMID: 39471860 Free PMC article. - A transposable element prevents severe hemophilia B and provides insights into the evolution of new- and old world primates.
Kopp J, Rovai A, Ott M, Wedemeyer H, Tiede A, Böhmer HJ, Marques T, Langemeier J, Bohne J, Krooss SA. Kopp J, et al. PLoS One. 2024 Oct 18;19(10):e0312303. doi: 10.1371/journal.pone.0312303. eCollection 2024. PLoS One. 2024. PMID: 39423215 Free PMC article. - Recognition of phylogenetically diverse pathogens through enzymatically amplified recruitment of RNF213.
Crespillo-Casado A, Pothukuchi P, Naydenova K, Yip MCJ, Young JM, Boulanger J, Dharamdasani V, Harper C, Hammoudi PM, Otten EG, Boyle K, Gogoi M, Malik HS, Randow F. Crespillo-Casado A, et al. EMBO Rep. 2024 Oct 7. doi: 10.1038/s44319-024-00280-w. Online ahead of print. EMBO Rep. 2024. PMID: 39375464 - Automated High-Throughput Biological Sex Identification from Archeological Human Dental Enamel Using Targeted Proteomics.
Koenig C, Bortel P, Paterson RS, Rendl B, Madupe PP, Troché GB, Hermann NV, Martínez de Pinillos M, Martinón-Torres M, Mularczyk S, Schjellerup Jørkov ML, Gerner C, Kanz F, Martinez-Val A, Cappellini E, Olsen JV. Koenig C, et al. J Proteome Res. 2024 Nov 1;23(11):5107-5121. doi: 10.1021/acs.jproteome.4c00557. Epub 2024 Sep 26. J Proteome Res. 2024. PMID: 39324540 Free PMC article.
References
- Goodman M, Porter CA, Czelusniak J, Page SL, Schneider H, et al. Toward a phylogenetic classification of Primates based on DNA evidence complemented by fossil evidence. Molecular Phylogenetics and Evolution. 1998;9:585–598. - PubMed
- Groves CP. Washington, DC: Smithsonian Institution Press; 2001. Primate taxonomy.350
- Wilson DE, Reeder DM. Baltimore: Johns Hopkins University Press; 2005. Mammal species of the world : a taxonomic and geographic reference.2142
- Fabre PH, Rodrigues A, Douzery EJP. Patterns of macroevolution among Primates inferred from a supermatrix of mitochondrial and nuclear DNA. Molecular Phylogenetics and Evolution. 2009;53:808–825. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous