Molecular architecture and function of matrix adhesions - PubMed (original) (raw)
Review
Molecular architecture and function of matrix adhesions
Benjamin Geiger et al. Cold Spring Harb Perspect Biol. 2011.
Abstract
Cell adhesions mediate important bidirectional interactions between cells and the extracellular matrix. They provide an interactive interface between the extracellular chemical and physical environment and the cellular scaffolding and signaling machinery. This dynamic, reciprocal regulation of intracellular processes and the matrix is mediated by membrane receptors such as the integrins, as well as many other components that comprise the adhesome. Adhesome constituents assemble themselves into different types of cell adhesion structures that vary in molecular complexity and change over time. These cell adhesions play crucial roles in cell migration, proliferation, and determination of cell fate.
Figures
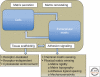
Figure 1.
Schematic illustration highlighting the dynamic cross talk between cells and the extracellular matrix (ECM). Cells secrete and remodel the ECM, and the ECM contributes to the assembly of individual cells into tissues, affecting this process at both receptor and cytoskeletal levels. Adhesion-mediated signaling, based on the cells’ capacity to sense the chemical and physical properties of the matrix, affects both global cell physiology and local molecular scaffolding of the adhesion sites. The molecular interactions within the adhesion site stimulate, in turn, the signaling process, by clustering together the structural and signaling components of the adhesome.
Figure 2.
Differential effects of different matrices on fibroblast spreading and FA formation. This figure shows morphological and molecular differences between integrin adhesions formed in response to adhesion to different ECM matrices. Vinculin (Vin)-labeled adhesions (green) are shown following adhesion of fibroblasts to fibronectin (FN) via the α5β1 integrin, or to vitronectin (VN) via the αvβ3 integrin. Notice that VN induces less cell spreading compared to cells adhering to FN, and that the distribution of FAs is largely peripheral. Original images for this panel were provided by Baruch Zimerman.
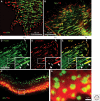
Figure 3.
Immunofluorescence images of different types of cell adhesions. (A) Fibronectin (FN) fibrils in human foreskin fibroblasts, despite their capacity to induce FA formation, are generally excluded from bona fide FAs; vinculin (red) and FN (green). (B) In contrast, FN fibrils are primarily associated with tensin-rich fibrillar adhesions; tensin (green) and FN (red). (C_–_E) Major forms of integrin adhesions formed by cultured porcine aortic endothelial cells and some of their molecular characteristics, in an endothelial cell labeled for paxillin (C: green) and tyrosine-phosphorylated paxillin (D: pY-paxillin, red), and the merged image (E). In these images, three major forms of integrin adhesions are detected: dotlike focal complexes (FX) located primarily at the cell’s leading edge, “classical” focal adhesions (FA), and fibrillar adhesions (FB) located near FAs but more toward the cell center, in which FN fibrils are prominent. Interestingly, the three types of cell adhesion differ in their molecular properties: In FX, paxillin is highly phosphorylated (about threefold higher than in FAs), whereas no paxillin phosphorylation is detected along FB (see white arrowheads). Additional molecular differences include, for example, the absence of zyxin from FXs, and selective enrichment of α5β1 and αvβ3 integrins in FB and FA, respectively. (F) and (G) show another form of integrin adhesion, podosomes, formed in this case by cultured osteoclasts. Podosomes consist of a core bundle of actin filaments (Act, green), oriented perpendicular to the plasma membrane; they are surrounded by a membrane-associated “adhesion ring” containing typical FA plaque molecules, including paxillin (red). In osteoclasts, podosomes accumulate along the cell edge, forming a belt-shaped “sealing zone” important for the process of bone resorption. The area in (F) marked with the white rectangle is enlarged in G, highlighting the relationship between the actin core and the adhesion zone. Original images for this figure were provided by Tova Volberg, Ronen Zaidel-Bar, and Chen Luxenburg.
Figure 4.
Scaffolding interactions of the integrin adhesome network (see
). Adhesome components include membrane receptors (dark green rectangles), adaptor proteins (purple rectangles), actin-associated proteins (magenta ovals), tyrosine kinases (red diamonds) and phosphatases (blue diamonds), serine/threonine kinases (red elongated hexagons) and phosphatases (blue elongated hexagons), G-proteins (orange ovals), GEFs (yellow diamonds), and GAPs (yellow elongated octagons). This diagram was prepared by Ronen Zaidel-Bar, based on Zaidel-Bar et al. 2007a; Zaidel-Bar and Geiger 2010.
Figure 5.
Regulatory (signaling) interactions of the integrin adhesome network; for details, see
. Signaling components include kinases, phosphatases, G-proteins and their regulators, as well as proteases. Red arrows point to modifications such as phosphorylation and activation of Rho GTPases, and blue arrows indicate dephosphorylation, inactivation of Rho GTPases, and protein degradation. The classes of molecules are indicated in the legend to Figure 4. This diagram was prepared by Ronen Zaidel-Bar, based on Zaidel-Bar et al. 2007a; Zaidel-Bar and Geiger 2010.
Figure 6.
Different views of FA structure: (A) Chicken lens cells were cultured on a flat surface, processed for transmission electron microscopy, and sectioned perpendicular to the plane of the substrate. The image reveals multiple cytoskeletal filaments accumulating at the adhesion site. The apparent gap between the ventral cell membrane and the substrate (indicated by a “serum line”) is approximately 10–15 nm. Bar = 1 µm. Original image provided by Ilana Sabanay. (B) Interference-reflection microscopy (IRM) of cell–matrix adhesions. This image shows cell adhesions of primary human fibroblasts, in which FAs appear dark gray or black, and the less tightly adhering “close contacts” are light gray. Bar = 10 µm. (Image adapted from Akiyama et al. 1989 and reprinted with permission from The Rockefeller University Press © 1989.) (C–F) Correlated microscopy, combining fluorescence microscopy (C) of FAs in fibroblasts expressing fluorescent paxillin to identify adhesion sites, and cryo-electron tomography (D–F). The inset in (C) shows a low-power image of the region bracketed on the fluorescence image. (D) A 10-nm slice through a cryo-electron tomogram of the focal adhesion indicated in the inset in (C), showing aligned actin filaments (white arrow), vesicle (black arrow), and the plasma membrane (white arrowhead). (E) Surface-rendering view of the focal adhesion site as seen from the direction of the substrate toward the cell. Actin is depicted in brown and membranes in blue; a large number of uniformly oriented particles, probably adhesion-related and depicted in green, are located at the interface between the cytoskeletal bundle and the membrane. No scale bar is shown in E, as it is a “rendered” version of D; the diameter of the enlarged “doughnut” shown in panel F is noted. (F) An enlarged view of an individual particle (diameter=25 nm), and the associated filaments. Panels C–F are based on data from Patla et al. 2010.
Similar articles
- Cell-matrix adhesion.
Berrier AL, Yamada KM. Berrier AL, et al. J Cell Physiol. 2007 Dec;213(3):565-73. doi: 10.1002/jcp.21237. J Cell Physiol. 2007. PMID: 17680633 Review. - Dimensions and dynamics in integrin function.
Yamada KM, Pankov R, Cukierman E. Yamada KM, et al. Braz J Med Biol Res. 2003 Aug;36(8):959-66. doi: 10.1590/s0100-879x2003000800001. Epub 2003 Jul 23. Braz J Med Biol Res. 2003. PMID: 12886449 Review. - Single and collective cell migration: the mechanics of adhesions.
De Pascalis C, Etienne-Manneville S. De Pascalis C, et al. Mol Biol Cell. 2017 Jul 7;28(14):1833-1846. doi: 10.1091/mbc.E17-03-0134. Mol Biol Cell. 2017. PMID: 28684609 Free PMC article. - The integrin adhesome: from genes and proteins to human disease.
Winograd-Katz SE, Fässler R, Geiger B, Legate KR. Winograd-Katz SE, et al. Nat Rev Mol Cell Biol. 2014 Apr;15(4):273-88. doi: 10.1038/nrm3769. Nat Rev Mol Cell Biol. 2014. PMID: 24651544 Review. - Analyzing the anatomy of integrin adhesions.
Byron A. Byron A. Sci Signal. 2011 Apr 26;4(170):jc3. doi: 10.1126/scisignal.2001896. Sci Signal. 2011. PMID: 21521876 Review.
Cited by
- ARAP2 signals through Arf6 and Rac1 to control focal adhesion morphology.
Chen PW, Jian X, Yoon HY, Randazzo PA. Chen PW, et al. J Biol Chem. 2013 Feb 22;288(8):5849-60. doi: 10.1074/jbc.M112.415778. Epub 2013 Jan 7. J Biol Chem. 2013. PMID: 23295182 Free PMC article. - Actin dynamics during tumor cell dissemination.
Mondal C, Di Martino JS, Bravo-Cordero JJ. Mondal C, et al. Int Rev Cell Mol Biol. 2021;360:65-98. doi: 10.1016/bs.ircmb.2020.09.004. Epub 2020 Nov 24. Int Rev Cell Mol Biol. 2021. PMID: 33962751 Free PMC article. Review. - Automated analysis of cell-matrix adhesions in 2D and 3D environments.
Broussard JA, Diggins NL, Hummel S, Georgescu W, Quaranta V, Webb DJ. Broussard JA, et al. Sci Rep. 2015 Jan 29;5:8124. doi: 10.1038/srep08124. Sci Rep. 2015. PMID: 25630460 Free PMC article. - Postnatal Pancreas of Mice Contains Tripotent Progenitors Capable of Giving Rise to Duct, Acinar, and Endocrine Cells In Vitro.
Ghazalli N, Mahdavi A, Feng T, Jin L, Kozlowski MT, Hsu J, Riggs AD, Tirrell DA, Ku HT. Ghazalli N, et al. Stem Cells Dev. 2015 Sep 1;24(17):1995-2008. doi: 10.1089/scd.2015.0007. Epub 2015 Jun 9. Stem Cells Dev. 2015. PMID: 25941840 Free PMC article. - Parvin-ILK: An intimate relationship.
Vakaloglou K, Zervas C. Vakaloglou K, et al. Bioarchitecture. 2012 May 1;2(3):91-94. doi: 10.4161/bioa.20700. Bioarchitecture. 2012. PMID: 22880148 Free PMC article.
References
- Abercrombie M, Heaysman JE, Pegrum SM 1970. The locomotion of fibroblasts in culture. I. Movements of the leading edge. Exp Cell Res 59: 393–398 - PubMed
- Alberts B, Johnson A, Lewis J, Raff M, Roberts K, Walter P 2008. Molecular biology of the cell. Garland Science, New York
Publication types
MeSH terms
Substances
Grants and funding
- PN2 EY016586/EY/NEI NIH HHS/United States
- U54GM64346/GM/NIGMS NIH HHS/United States
- ZIA DE000524-21/Intramural NIH HHS/United States
- U54 GM064346/GM/NIGMS NIH HHS/United States
- ZIA DE000718-05/Intramural NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources