RNA in defense: CRISPRs protect prokaryotes against mobile genetic elements - PubMed (original) (raw)
RNA in defense: CRISPRs protect prokaryotes against mobile genetic elements
Matthijs M Jore et al. Cold Spring Harb Perspect Biol. 2012.
Abstract
The CRISPR/Cas system in prokaryotes provides resistance against invading viruses and plasmids. Three distinct stages in the mechanism can be recognized. Initially, fragments of invader DNA are integrated as new spacers into the repetitive CRISPR locus. Subsequently, the CRISPR is transcribed and the transcript is cleaved by a Cas protein within the repeats, generating short RNAs (crRNAs) that contain the spacer sequence. Finally, crRNAs guide the Cas protein machinery to a complementary invader target, either DNA or RNA, resulting in inhibition of virus or plasmid proliferation. In this article, we discuss our current understanding of this fascinating adaptive and heritable defense system, and describe functional similarities and differences with RNAi in eukaryotes.
Figures
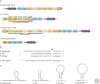
Figure 1.
Overview of the four CRISPR/Cas subtypes that are described in this article. For an overview of all eight CRISPR/Cas subtypes, see Haft et al. 2005 and van der Oost et al. 2009. (A) cas gene neighborhoods in four experimentally studied organisms, each representing a different subtype indicated between brackets. CRISPRs consist of a leader (grey box), repeats (red diamonds), and spacers (blue boxes). Only a fragment of the CRISPR is shown. Genes are indicated as arrows. Blue arrows indicate genes that are (possibly) involved in spacer acquisition. Yellow arrows indicate genes that are involved in CRISPR transcription and processing and target interference. The endonucleases that cleave pre-crRNA generating crRNA are highlighted as bold arrows. Hatching patterns indicate gene similarity: RAMP genes have vertical lines, polymerase genes have horizontal lines, CasC homologs have diagonal lines, and other genes that are not related to each other are filled. Genes that encode proteins from isolated complexes (Cse-complex from E. coli and Cmr-complex from Pyrococcus furiosus) are underlined. (B) CRISPR RNA repeat sequences from each organism are given. The cleavage site is indicated by a triangle. Although the repeat sequences are different, all CRISPR RNA cleavage events generate an eight-nucleotide 5′ handle. Please note that the cleavage site in Streptococcus thermophilus CRISPR RNA has not been determined. Palindromic sequences are underlined. (C) Predicted secondary structures of the different CRISPR RNA repeats. Cleavage sites are indicated with an arrow. As described previously by Kunin et al., the repeat of P. furiosus is not likely to form a stem loop (Kunin et al. 2007).
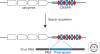
Figure 2.
Integration of a new spacer. A new spacer is acquired at the leader proximal side of the CRISPR during virus infection, resulting in resistance. The CRISPR consist of a leader (grey box), repeats (red diamonds), and spacers (blue boxes). The newly acquired spacer is numbered 0 and matches the sequence of the virus (proto-spacer). The proto-spacer adjacent motif (PAM) is located downstream or upstream the proto-spacer.
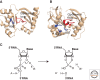
Figure 3.
The catalytic sites of CasE and Cas6, and the proposed reaction mechanism of pre-crRNA cleavage. (A) Proposed catalytic site of CasE from T. thermophilus showing the conserved histidine residue (H26) and the glycine-rich carboxy-terminal loop. The catalytic site of Cas6 from P. furiosus (B) contains a catalytic triad of tyrosine (Y31), histidine (H46) and lysine (K52) and a glycine-rich carboxy-terminal loop. The loop and the overall duplicated ferredoxin fold are conserved among CasE and Cas6. Pre-crRNA cleavage might follow a general acid–base hydrolysis mechanism (C). A base (B) draws a proton from the 2′OH of the ribose ring. A subsequent nucleophilic attack on the phosphorus atom is simultaneously compensated by the acid (A) that donates a proton to the leaving 3′RNA. The tyrosine residue of Cas6 is proposed to be the base and the histidine the acid residue (Carte et al. 2008). In CasE the histidine and a water molecule might be the catalytic residues. Pictures in (A) and (B) are generated with pymol (
), potential catalytic residues are depicted in blue; the glycine-rich loop is depicted in red. Coordinates were obtained from the Protein Data Bank (
).
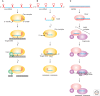
Figure 4.
Antiviral DNA and RNA silencing pathways in prokaryotes and eukaryotes. (A) crRNA mediated DNA silencing pathway in E. coli. pre-crRNA is cleaved by the CasE subunit of Cascade (Cse-complex) and the mature crRNA remains bound to Cascade. How Cascade, assisted by Cas3, recognizes and neutralizes invading DNA remains to be elucidated. (B) crRNA mediated RNA silencing pathway in P. furiosus. Pre-crRNA is cleaved by Cas6 and then further trimmed to generate crRNAs of two different lengths. These crRNAs are bound by the Cmr-complex. This loaded Cmr-complex specifically binds viral RNA and cleaves the complementary strand 14 nucleotides away from the 3′ end of the crRNA. This pathway shares functional analogies with siRNA mediated antiviral resistance in eukaryotes. (C) siRNAs are generated from viral dsRNA by dicer. The first (random) cleavage event by dicer generates dsRNA with a 3′ dinucleotide overhang. The second cleavage by dicer takes place 20–25 bases away from the overhang, generating short dsRNAs. The dsRNA is transferred to the Argonaute protein of the RISC complex and the passenger strand is removed. The retained guide strand can basepair with a complementary viral mRNA molecule, followed by a cleavage of the scissile bond between the 10th and 11th base from the 3′ end of the guide strand. The cleaved target RNA dissociates and the recycled RISC can be used in a second round of RNA binding and cleavage. Please note that dashed arrows indicate processes that are based on hypotheses.
Similar articles
- Clustered regularly interspaced short palindromic repeats (CRISPRs): the hallmark of an ingenious antiviral defense mechanism in prokaryotes.
Al-Attar S, Westra ER, van der Oost J, Brouns SJ. Al-Attar S, et al. Biol Chem. 2011 Apr;392(4):277-89. doi: 10.1515/BC.2011.042. Epub 2011 Feb 7. Biol Chem. 2011. PMID: 21294681 Review. - CRISPR-based adaptive and heritable immunity in prokaryotes.
van der Oost J, Jore MM, Westra ER, Lundgren M, Brouns SJ. van der Oost J, et al. Trends Biochem Sci. 2009 Aug;34(8):401-7. doi: 10.1016/j.tibs.2009.05.002. Epub 2009 Jul 29. Trends Biochem Sci. 2009. PMID: 19646880 Review. - Prokaryotic homologs of Argonaute proteins are predicted to function as key components of a novel system of defense against mobile genetic elements.
Makarova KS, Wolf YI, van der Oost J, Koonin EV. Makarova KS, et al. Biol Direct. 2009 Aug 25;4:29. doi: 10.1186/1745-6150-4-29. Biol Direct. 2009. PMID: 19706170 Free PMC article. - Substrate generation for endonucleases of CRISPR/cas systems.
Zoephel J, Dwarakanath S, Richter H, Plagens A, Randau L. Zoephel J, et al. J Vis Exp. 2012 Sep 8;(67):4277. doi: 10.3791/4277. J Vis Exp. 2012. PMID: 22986408 Free PMC article. - CRISPR-Cas systems and RNA-guided interference.
Barrangou R. Barrangou R. Wiley Interdiscip Rev RNA. 2013 May-Jun;4(3):267-78. doi: 10.1002/wrna.1159. Epub 2013 Mar 20. Wiley Interdiscip Rev RNA. 2013. PMID: 23520078 Review.
Cited by
- Viral vectors and extracellular vesicles: innate delivery systems utilized in CRISPR/Cas-mediated cancer therapy.
Ahmadi SE, Soleymani M, Shahriyary F, Amirzargar MR, Ofoghi M, Fattahi MD, Safa M. Ahmadi SE, et al. Cancer Gene Ther. 2023 Jul;30(7):936-954. doi: 10.1038/s41417-023-00597-z. Epub 2023 Feb 28. Cancer Gene Ther. 2023. PMID: 36854897 Free PMC article. Review. - Cascade-mediated binding and bending of negatively supercoiled DNA.
Westra ER, Nilges B, van Erp PB, van der Oost J, Dame RT, Brouns SJ. Westra ER, et al. RNA Biol. 2012 Sep;9(9):1134-8. doi: 10.4161/rna.21410. Epub 2012 Sep 1. RNA Biol. 2012. PMID: 22954644 Free PMC article. Review. - Mechanisms of Evolutionary Innovation Point to Genetic Control Logic as the Key Difference Between Prokaryotes and Eukaryotes.
Bains W, Schulze-Makuch D. Bains W, et al. J Mol Evol. 2015 Aug;81(1-2):34-53. doi: 10.1007/s00239-015-9688-6. Epub 2015 Jul 25. J Mol Evol. 2015. PMID: 26208881 - Novel CRISPR-Cas Systems: An Updated Review of the Current Achievements, Applications, and Future Research Perspectives.
Nidhi S, Anand U, Oleksak P, Tripathi P, Lal JA, Thomas G, Kuca K, Tripathi V. Nidhi S, et al. Int J Mol Sci. 2021 Mar 24;22(7):3327. doi: 10.3390/ijms22073327. Int J Mol Sci. 2021. PMID: 33805113 Free PMC article. Review. - Unusual genome complexity in Lactobacillus salivarius JCM1046.
Raftis EJ, Forde BM, Claesson MJ, O'Toole PW. Raftis EJ, et al. BMC Genomics. 2014 Sep 8;15(1):771. doi: 10.1186/1471-2164-15-771. BMC Genomics. 2014. PMID: 25201645 Free PMC article.
References
- Agari Y, Yokoyama S, Kuramitsu S, Shinkai A 2008. X-ray crystal structure of a CRISPR-associated protein, Cse2, from Thermus thermophilus HB8. Proteins 73: 1063–1067 - PubMed
- Andersson AF, Banfield JF 2008. Virus population dynamics and acquired virus resistance in natural microbial communities. Science 320: 1047–1050 - PubMed
- Aravind L, Koonin EV 1998. The HD domain defines a new superfamily of metal-dependent phosphohydrolases. Trends Biochem Sci 23: 469–472 - PubMed
- Banfield JF, Young M 2009. Microbiology. Variety–the splice of life–in microbial communities. Science 326: 1198–1199 - PubMed
- Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P 2007. CRISPR provides acquired resistance against viruses in prokaryotes. Science 315: 1709–1712 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources