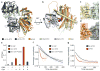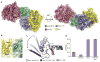A conserved mechanism of DEAD-box ATPase activation by nucleoporins and InsP6 in mRNA export - PubMed (original) (raw)
. 2011 Apr 14;472(7342):238-42.
doi: 10.1038/nature09862. Epub 2011 Mar 27.
Affiliations
- PMID: 21441902
- PMCID: PMC3078754
- DOI: 10.1038/nature09862
A conserved mechanism of DEAD-box ATPase activation by nucleoporins and InsP6 in mRNA export
Ben Montpetit et al. Nature. 2011.
Abstract
Superfamily 1 and superfamily 2 RNA helicases are ubiquitous messenger-RNA-protein complex (mRNP) remodelling enzymes that have critical roles in all aspects of RNA metabolism. The superfamily 2 DEAD-box ATPase Dbp5 (human DDX19) functions in mRNA export and is thought to remodel mRNPs at the nuclear pore complex (NPC). Dbp5 is localized to the NPC via an interaction with Nup159 (NUP214 in vertebrates) and is locally activated there by Gle1 together with the small-molecule inositol hexakisphosphate (InsP(6)). Local activation of Dbp5 at the NPC by Gle1 is essential for mRNA export in vivo; however, the mechanistic role of Dbp5 in mRNP export is poorly understood and it is not known how Gle1(InsP6) and Nup159 regulate the activity of Dbp5. Here we report, from yeast, structures of Dbp5 in complex with Gle1(InsP6), Nup159/Gle1(InsP6) and RNA. These structures reveal that InsP(6) functions as a small-molecule tether for the Gle1-Dbp5 interaction. Surprisingly, the Gle1(InsP6)-Dbp5 complex is structurally similar to another DEAD-box ATPase complex essential for translation initiation, eIF4G-eIF4A, and we demonstrate that Gle1(InsP6) and eIF4G both activate their DEAD-box partner by stimulating RNA release. Furthermore, Gle1(InsP6) relieves Dbp5 autoregulation and cooperates with Nup159 in stabilizing an open Dbp5 intermediate that precludes RNA binding. These findings explain how Gle1(InsP6), Nup159 and Dbp5 collaborate in mRNA export and provide a general mechanism for DEAD-box ATPase regulation by Gle1/eIF4G-like activators.
Conflict of interest statement
The authors declare no competing financial interests.
Figures
Figure 1. The Gle1IP6-Δ90Dbp5-ADP complex
a, Structure of Dbp5 bound to ADP (magenta spheres) and Gle1 (see colour key). b, Side view showing IP6 (coloured spheres) bound at the interface between Gle1 and Dbp5. c, Detailed view of the IP6 binding interface (see colour key). IP6 is shown as grey sticks with orange phosphate and red oxygen atoms. Nitrogen atoms are in dark blue. d, IP6 binding is required for maximal Dbp5 ATPase stimulation with RNA. Error bars represent s.d. (n=3). e, Structural superposition of the RNA-Δ90Dbp5 complex with Gle1IP6-Δ90Dbp5. Arrows depict large rigid body movement in Dbp5. f, Van der Waals surface view of the RNA-Δ90Dbp5 complex and the Gle1IP6-Δ90Dbp5 complex coloured by solvent accessible electrostatics. Abbreviations: kb – Boltzmann’s constant (Joules/Kelvin), T – temperature (310 Kelvin), ec – charge of an electron (1.602 x 10−19 Coulombs).
Figure 2. Comparison of Gle1IP6-Δ90Dbp5 and eIF4A-eIF4G
a, Structural superposition of Gle1IP6-Δ90Dbp5 with eIF4G-eIF4A and AMP (PDB ID: 2VSO) (see colour key). b, View of the C-terminal RecA-like domain binding interface. Unique α-helices present in both Dbp5 and Gle1 form the IP6 biding pocket (boxed in figure). c, d, Residues involved in the formation of the N-terminal RecA-like domain binding interface in (c) Gle1IP6-Δ90Dbp5 and (d) eIF4A-eIF4G. e, Measured ATPase activity using wild type or mutant Gle1. Error bars represent s.d. (n=3). f, g, RNA release from Dbp5E240Q or eIF4AE172Q monitored by fluorescence polarization. Representative curves are shown.
Figure 3. The Gle1IP6-Δ90Dbp5-Nup159 complex
a, Two views of the Gle1IP6-Dbp5-Nup159 complex (see colour key). b, The N-terminal RecA-like domain binding interface between Dbp5- Gle1IP6 is altered in the presence of Nup159 (compare to Fig. 2c). Residues involved in the binding interface (sticks) are labelled. c, Superposition of the N-terminal RecA-domain among the three structural states of Dbp5. Arrow highlights the movement of the CTD and catalytic arginine finger residues (sticks) among the three states (see colour key). For clarity, the Gle1-ADP intermediate is shown slightly transparent, and both the arginine finger side chains and ADP have been removed. d, Inhibition of the RNA stimulated ATPase activity of Dbp5 by Nup159 is overcome in the presence of Gle1. Error bars represent s.d. (n=3).
Figure 4. Model of the Dbp5 mechanochemical cycle
In the presence ATP, Dbp5 binds RNA causing local destabilization and remodelling of duplexed RNA or RNA-protein complexes (state I). ATP hydrolysis then allows the activator (Gle1) to bind both the C-terminal and N-terminal RecA-like domains, separating the two RecA-like domains and promoting RNA release (state II). For Dbp5 it is currently not known when Pi is released following hydrolysis, but this will likely occur prior to the formation of state II. Subsequent release of the bound RNA allows Nup159 to bind Dbp5 causing the two RecA domains to further separate (state III). The formation of this state could then facilitate ADP release, prevent rebinding of the RNA, and aid in enzyme recycling (state IV). Crystal structure model in state IV is PDB ID:3FHO. For colour coding details see key.
Similar articles
- Inositol hexakisphosphate and Gle1 activate the DEAD-box protein Dbp5 for nuclear mRNA export.
Alcázar-Román AR, Tran EJ, Guo S, Wente SR. Alcázar-Román AR, et al. Nat Cell Biol. 2006 Jul;8(7):711-6. doi: 10.1038/ncb1427. Epub 2006 Jun 18. Nat Cell Biol. 2006. PMID: 16783363 - Activation of the DExD/H-box protein Dbp5 by the nuclear-pore protein Gle1 and its coactivator InsP6 is required for mRNA export.
Weirich CS, Erzberger JP, Flick JS, Berger JM, Thorner J, Weis K. Weirich CS, et al. Nat Cell Biol. 2006 Jul;8(7):668-76. doi: 10.1038/ncb1424. Epub 2006 Jun 18. Nat Cell Biol. 2006. PMID: 16783364 - The Dbp5 cycle at the nuclear pore complex during mRNA export I: dbp5 mutants with defects in RNA binding and ATP hydrolysis define key steps for Nup159 and Gle1.
Hodge CA, Tran EJ, Noble KN, Alcazar-Roman AR, Ben-Yishay R, Scarcelli JJ, Folkmann AW, Shav-Tal Y, Wente SR, Cole CN. Hodge CA, et al. Genes Dev. 2011 May 15;25(10):1052-64. doi: 10.1101/gad.2041611. Genes Dev. 2011. PMID: 21576265 Free PMC article. - Dbp5, Gle1-IP6 and Nup159: a working model for mRNP export.
Folkmann AW, Noble KN, Cole CN, Wente SR. Folkmann AW, et al. Nucleus. 2011 Nov-Dec;2(6):540-8. doi: 10.4161/nucl.2.6.17881. Epub 2011 Nov 1. Nucleus. 2011. PMID: 22064466 Free PMC article. Review. - Dbp5 - from nuclear export to translation.
Tieg B, Krebber H. Tieg B, et al. Biochim Biophys Acta. 2013 Aug;1829(8):791-8. doi: 10.1016/j.bbagrm.2012.10.010. Epub 2012 Nov 2. Biochim Biophys Acta. 2013. PMID: 23128325 Review.
Cited by
- DEAD-box helicases as integrators of RNA, nucleotide and protein binding.
Putnam AA, Jankowsky E. Putnam AA, et al. Biochim Biophys Acta. 2013 Aug;1829(8):884-93. doi: 10.1016/j.bbagrm.2013.02.002. Epub 2013 Feb 15. Biochim Biophys Acta. 2013. PMID: 23416748 Free PMC article. Review. - Crystal structure of the N-terminal domain of Nup358/RanBP2.
Kassube SA, Stuwe T, Lin DH, Antonuk CD, Napetschnig J, Blobel G, Hoelz A. Kassube SA, et al. J Mol Biol. 2012 Nov 9;423(5):752-65. doi: 10.1016/j.jmb.2012.08.026. Epub 2012 Sep 7. J Mol Biol. 2012. PMID: 22959972 Free PMC article. - Dbp5 associates with RNA-bound Mex67 and Nab2 and its localization at the nuclear pore complex is sufficient for mRNP export and cell viability.
Adams RL, Wente SR. Adams RL, et al. PLoS Genet. 2020 Oct 1;16(10):e1009033. doi: 10.1371/journal.pgen.1009033. eCollection 2020 Oct. PLoS Genet. 2020. PMID: 33002012 Free PMC article. - The nucleoporin Gle1 activates DEAD-box protein 5 (Dbp5) by promoting ATP binding and accelerating rate limiting phosphate release.
Gray S, Cao W, Montpetit B, De La Cruz EM. Gray S, et al. Nucleic Acids Res. 2022 Apr 22;50(7):3998-4011. doi: 10.1093/nar/gkac164. Nucleic Acids Res. 2022. PMID: 35286399 Free PMC article. - Structural basis for RNA-duplex unwinding by the DEAD-box helicase DbpA.
Wurm JP. Wurm JP. RNA. 2023 Sep;29(9):1339-1354. doi: 10.1261/rna.079582.123. Epub 2023 May 23. RNA. 2023. PMID: 37221012 Free PMC article.
References
- Cordin O, Banroques J, Tanner NK, Linder P. The DEAD-box protein family of RNA helicases. Gene. 2006;367:17–37. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM058065/GM/NIGMS NIH HHS/United States
- T32 GM007232/GM/NIGMS NIH HHS/United States
- R00 GM080097/GM/NIGMS NIH HHS/United States
- RC1GM91533/GM/NIGMS NIH HHS/United States
- RC1 GM091533/GM/NIGMS NIH HHS/United States
- R01GM58065/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous