Crystal structure of a coiled-coil domain from human ROCK I - PubMed (original) (raw)
Crystal structure of a coiled-coil domain from human ROCK I
Daqi Tu et al. PLoS One. 2011.
Abstract
The small GTPase Rho and one of its targets, Rho-associated kinase (ROCK), participate in a variety of actin-based cellular processes including smooth muscle contraction, cell migration, and stress fiber formation. The ROCK protein consists of an N-terminal kinase domain, a central coiled-coil domain containing a Rho binding site, and a C-terminal pleckstrin homology domain. Here we present the crystal structure of a large section of the central coiled-coil domain of human ROCK I (amino acids 535-700). The structure forms a parallel α-helical coiled-coil dimer that is structurally similar to tropomyosin, an actin filament binding protein. There is an unusual discontinuity in the coiled-coil; three charged residues (E613, R617 and D620) are positioned at what is normally the hydrophobic core of coiled-coil packing. We speculate that this conserved irregularity could function as a hinge that allows ROCK to adopt its autoinhibited conformation.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
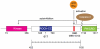
Figure 1. Schematic view of the ROCK I domain structure.
In the inactive form, the Rho-binding domain (RBD) and the pleckstrin homology (PH) domain bind to the amino-terminal kinase domain. This autoinhibitory interaction is released either by GTP-Rho binding to RBD, or by caspase-3 cleavage of the PH domain. PH domain contains an internal cysteine-rich domain (CRD). The entire central domain (422–1102) is predicted to be coiled-coil, as indicated. ROCK-CC (in blue) is the solved crystal structure region in this report. The position of residue 617 is indicated with an arrow.
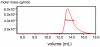
Figure 2. ROCK-CC forms a dimer in solution.
Purified ROCK (535–704) was analyzed on a Superdex 200 gel filtration column coupled to a multi-angle light scattering detector. The protein elution profile measured by refractive index is shown as a thin red trace; the horizontal thick red line is measured molar mass (∼40.5 KDa). Theoretical molar mass of a dimer is 40.4 KDa.
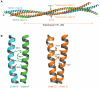
Figure 3. Overall structure of ROCK-CC.
(A) Ribbon diagram of the ROCK-CC dimer is shown with chain A in green and chain B in cyan. The parallel coiled-coil homo-dimer is asymmetric. (B) Crystal packing of dimer A/B with dimer C/D. The orientation is that of Fig. 3A turned 90° towards the viewer along the superhelical axis. Chain C is colored in yellow, chain D is in magenta.
Figure 4. Inter-helical interactions in ROCK-CC.
(A) In sphere representation, knobs from chain A (green) pack into holes from chain B (cyan). For clarity, only side chains are shown and knobs from chain B packing into holes from chain A are not displayed. (B) Helical wheel diagram of residues Asn-589 to Glu-665. Interhelical polar interactions are indicated by a line connecting paired residues. E613 and D620 at the d positions are colored red. R617 at the a position is colored blue. (C) Inter-Helical distance (HD) for two structures. Due to crystal packing, only residues 589 to 666 assume coiled-coil conformation for dimer A/B; for dimer C/D, residues 578 to 666 are in a coiled-coil conformation. Note that both dimers have a peak in their inter-helical distance at the E613/R617/D620 junction. (D) Average B-factors for mainchain atoms at each residue for all four chains. Since the structure was refined using TLS, the B-factor plotted represents the isotropic equivalent of the total B-factor (B_tls + B_individual).
Figure 5. Stereogram of electron density map at the coiled-coil interface in the region of the E613/R617/D620 bulge.
Chain C is shown in yellow and chain D in grey. The σA weighted 2FO – FC map is contoured at 1.5 σ and computed anisotropically to 2.3 Å. Due to the anisotropy, the map was sharpened by applying a B factor scaling of -10 Å2 (see methods).
Figure 6. Superposition of ROCK-CC and tropomyosin (PDB code 2d3e).
(A) DALI alignment matrix was based on only aligning ROCK-CC chain A to chain C in the tropomyosin structure. The tropomyosin structure is a fusion with the GCN4 leucine zipper; the tropomyosin portion of the structure (residues 176–282) is indicated below the superposition. Positions of R617 of ROCK chain A and E218 of tropomyosin chain D are indicated by arrows. (B) Detailed views of the respective unusual junctions in ROCK (left) and tropomyosin (right).
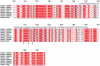
Figure 7. Sequence alignment of the ROCK coiled-coil region.
Residues 535–700 is the region used for this crystallographic study. Identical residues are indicated by the red background, similar residues by red characters. The experimentally identified _a_-g heptad repeats are displayed above the sequence for residues 589 to 665. Residues E613, R617 and D620 are marked by arrows.
Similar articles
- Structure of a highly conserved domain of Rock1 required for Shroom-mediated regulation of cell morphology.
Mohan S, Das D, Bauer RJ, Heroux A, Zalewski JK, Heber S, Dosunmu-Ogunbi AM, Trakselis MA, Hildebrand JD, Vandemark AP. Mohan S, et al. PLoS One. 2013 Dec 9;8(12):e81075. doi: 10.1371/journal.pone.0081075. eCollection 2013. PLoS One. 2013. PMID: 24349032 Free PMC article. - Parallel coiled-coil association of the RhoA-binding domain in Rho-kinase.
Shimizu T, Ihara K, Maesaki R, Amano M, Kaibuchi K, Hakoshima T. Shimizu T, et al. J Biol Chem. 2003 Nov 14;278(46):46046-51. doi: 10.1074/jbc.M306458200. Epub 2003 Sep 3. J Biol Chem. 2003. PMID: 12954645 - N-terminus-mediated dimerization of ROCK-I is required for RhoE binding and actin reorganization.
Garg R, Riento K, Keep N, Morris JD, Ridley AJ. Garg R, et al. Biochem J. 2008 Apr 15;411(2):407-14. doi: 10.1042/BJ20071342. Biochem J. 2008. PMID: 18215121 - Rho-associated coiled-coil containing kinases (ROCK): structure, regulation, and functions.
Julian L, Olson MF. Julian L, et al. Small GTPases. 2014;5:e29846. doi: 10.4161/sgtp.29846. Epub 2014 Jul 10. Small GTPases. 2014. PMID: 25010901 Free PMC article. Review. - Tropomyosin Structure, Function, and Interactions: A Dynamic Regulator.
Hitchcock-DeGregori SE, Barua B. Hitchcock-DeGregori SE, et al. Subcell Biochem. 2017;82:253-284. doi: 10.1007/978-3-319-49674-0_9. Subcell Biochem. 2017. PMID: 28101865 Review.
Cited by
- Structural basis for recognition of autophagic receptor NDP52 by the sugar receptor galectin-8.
Kim BW, Hong SB, Kim JH, Kwon DH, Song HK. Kim BW, et al. Nat Commun. 2013;4:1613. doi: 10.1038/ncomms2606. Nat Commun. 2013. PMID: 23511477 - Structure of Shroom domain 2 reveals a three-segmented coiled-coil required for dimerization, Rock binding, and apical constriction.
Mohan S, Rizaldy R, Das D, Bauer RJ, Heroux A, Trakselis MA, Hildebrand JD, VanDemark AP. Mohan S, et al. Mol Biol Cell. 2012 Jun;23(11):2131-42. doi: 10.1091/mbc.E11-11-0937. Epub 2012 Apr 4. Mol Biol Cell. 2012. PMID: 22493320 Free PMC article. - Structure of the Shroom-Rho Kinase Complex Reveals a Binding Interface with Monomeric Shroom That Regulates Cell Morphology and Stimulates Kinase Activity.
Zalewski JK, Mo JH, Heber S, Heroux A, Gardner RG, Hildebrand JD, VanDemark AP. Zalewski JK, et al. J Biol Chem. 2016 Dec 2;291(49):25364-25374. doi: 10.1074/jbc.M116.738559. Epub 2016 Oct 10. J Biol Chem. 2016. PMID: 27758857 Free PMC article. - Structure of a highly conserved domain of Rock1 required for Shroom-mediated regulation of cell morphology.
Mohan S, Das D, Bauer RJ, Heroux A, Zalewski JK, Heber S, Dosunmu-Ogunbi AM, Trakselis MA, Hildebrand JD, Vandemark AP. Mohan S, et al. PLoS One. 2013 Dec 9;8(12):e81075. doi: 10.1371/journal.pone.0081075. eCollection 2013. PLoS One. 2013. PMID: 24349032 Free PMC article. - Dietary Calcium Alleviates Fluorine-Induced Liver Injury in Rats by Mitochondrial Apoptosis Pathway.
Li H, Hao Z, Wang L, Yang J, Zhao Y, Cheng X, Yuan H, Wang J. Li H, et al. Biol Trace Elem Res. 2022 Jan;200(1):271-280. doi: 10.1007/s12011-021-02641-1. Epub 2021 Feb 24. Biol Trace Elem Res. 2022. PMID: 33629228
References
- Nakagawa O, Fujisawa K, Ishizaki T, Saito Y, Nakao K, et al. ROCK-I and ROCK-II, two isoforms of Rho-associated coiled-coil forming protein serine/threonine kinase in mice. FEBS Lett. 1996;392:189–193. - PubMed
- Leung T, Manser E, Tan L, Lim L. A novel serine/threonine kinase binding the Ras-related RhoA GTPase which translocates the kinase to peripheral membranes. J Biol Chem. 1995;270:29051–29054. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases