Depletion of murine intestinal microbiota: effects on gut mucosa and epithelial gene expression - PubMed (original) (raw)
Depletion of murine intestinal microbiota: effects on gut mucosa and epithelial gene expression
Dag Henrik Reikvam et al. PLoS One. 2011.
Abstract
Background: Inappropriate cross talk between mammals and their gut microbiota may trigger intestinal inflammation and drive extra-intestinal immune-mediated diseases. Epithelial cells constitute the interface between gut microbiota and host tissue, and may regulate host responses to commensal enteric bacteria. Gnotobiotic animals represent a powerful approach to study bacterial-host interaction but are not readily accessible to the wide scientific community. We aimed at refining a protocol that in a robust manner would deplete the cultivable intestinal microbiota of conventionally raised mice and that would prove to have significant biologic validity.
Methodology/principal findings: Previously published protocols for depleting mice of their intestinal microbiota by administering broad-spectrum antibiotics in drinking water were difficult to reproduce. We show that twice daily delivery of antibiotics by gavage depleted mice of their cultivable fecal microbiota and reduced the fecal bacterial DNA load by 400 fold while ensuring the animals' health. Mice subjected to the protocol for 17 days displayed enlarged ceca, reduced Peyer's patches and small spleens. Antibiotic treatment significantly reduced the expression of antimicrobial factors to a level similar to that of germ-free mice and altered the expression of 517 genes in total in the colonic epithelium. Genes involved in cell cycle were significantly altered concomitant with reduced epithelial proliferative activity in situ assessed by Ki-67 expression, suggesting that commensal microbiota drives cellular proliferation in colonic epithelium.
Conclusion: We present a robust protocol for depleting conventionally raised mice of their cultivatable intestinal microbiota with antibiotics by gavage and show that the biological effect of this depletion phenocopies physiological characteristics of germ-free mice.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
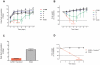
Figure 1. Mice refrain from drinking antibiotics ad libitum.
Experiments attempting to administer mice ampicillin 1 g/l (A), vancomycin 500 mg/ml (V), neomycin 1 g/l (N), and metronidazol 1 g/l (M) in drinking water ad libitum. Water indicates mice receiving regular drinking water. Skulls represent euthanized animals. Stapled lines indicate humane endpoint of 20% loss of body weight. Treatment groups presented as mean ± SD. Deceased mice excluded from subsequent time points after death. (A) Daily fluid consumption estimated by daily weighting of flasks and (B) bodyweight presented as percent of baseline ( = Day 0) for mice receiving the full AVNM concoction (red) in drinking water as well as for mice receiving the individual antibiotics as single solutions in drinking water, n = 5 for all groups. (C) Daily fluid consumption and (D) bodyweight presented as percent of baseline ( = Day 0) attempting to mask the foul taste of the antibiotic concoction AVNM in drinking water by adding the aspartame-based sweetener Canderel® 1.5% weight/volume, n = 6 for AVNM + Canderel® 1.5% group, n = 4 for water control group.
Figure 2. Protocol for administering antibiotics by gavage ensures mice health.
(A) Sketch of protocol for gavage administration of vancomycin 50 mg/kg (V), neomycin 100 mg/kg (N), metronidazol 100 mg/kg (M), and amphotericin-B 1 mg/kg (Amf-B) every 12 hours with ampicillin 1 g/l ad libitum in drinking water (A). (B) Body weight, presented as percent of base line ( = Day 0), of mice successfully depleted of cultivable fecal microbiota by gavage (VNMAmf-B+A) or gavaged with water in equivalent volume and frequency (Sham). Control mice received water ad libitum only
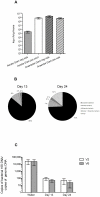
Figure 3. Protocol for administering antibiotics by gavage effectively reduces intestinal bacterial contents.
(A) Enumeration of cultivable fecal bacteria (mean ± SEM) in untreated mice. (B) Efficacy of successful depletion (<1 cfu/mg feces) of cultivable gut microbiota and distribution of aerobe, anaerobe, and fungal overgrowth (def.: ≥1 cfu/mg feces) after 13 and 24 days of treatment with vancomycinm (V), neomycin (N), metronidazol (M), and amphotericin-B (Amf-B) every 12 hours by gavage with ampicillin ad libitum in drinking water (A), n = 43. (C) Bacterial 16S DNA load (mean ± SD) in fecal pellets. 16S DNA determined by quantitative PCR for the V2 and V6 regions and normalized to total mouse genomic DNA in the same pellets of mice treated 13 and 24 days with VNMAmf-B +A compared with water controls, n = 7 for water controls, 40 for 13 days, and 35 for 24 days.
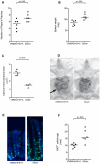
Figure 4. Mice subjected to antibiotic gavage treatment alters macroscopic phenotype and epithelial proliferative activity.
Numbers of macroscopically visible Peyer's patches (A), spleen weight (B), and cecal longitudinal cross section area (C), after 17 days of treatment with vancomycin (V), neomycin (N), metronidazol (M), and amphotericin-B (Amf-B) every 12 hours by gavage with ampicillin ad libitum in drinking water (A) compared to mice gavaged with water only (Sham). All VNMAmf-B+A treated mice exhibited successful depletion of cultivable intestinal microbiota (<1 cfu/mg feces). (D) Photograph of VNMAmf-B+A and sham fed mice. Note the enlarged cecum (arrow) of the antibiotic treated mouse. (E) Immunofluorescent staining for Ki67 (green) and DNA (Hoechst dye; blue) of cross sections of colons from VNMAmf-B+A fed mice and sham fed mice with (F) enumeration of Ki67+ cells per crypt. In all graphs each symbol represents one animal and horizontal bars represent medians. Statistical differences calculated by Mann-Whitney two-tailed test.
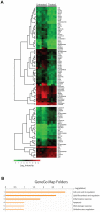
Figure 5. Antibiotic gavage treatment alters gene expression profile of the colonic epithelium.
Gene expression profile of colonic epithelial cells isolated from mice subjected to 17 days of treatment with vancomycin, neomycin, metronidazol, and amphotericin-B every 12 hours by gavage with ampicillin ad libitum in drinking water (Treated) compared to mice gavaged with water only (Untreated). All Treated mice exhibited successful depletion of cultivable intestinal microbiota (<1 cfu/mg feces). (**A**) Heat-map analysis of significantly (p<0.05) differentially expressed genes with >two-fold change organized in dendrogram according to Eukledian relation of the differentially expressed genes. Colors indicate absolute expression of one gene estimated by mean values of the multiple probes detecting each gene on the chip. Each row represent one gene, each column represent one mouse. (B) Gene ontology map folder enrichment analysis of differentially expressed genes in MetaCore (GeneGo, St Joseph, MI).
Figure 6. Quantitative PCR validates colonic IEC gene expression profiles of antibiotic treatment in relation to germ-free mice.
Quantitative reverse transcriptase PCR of colonic intestinal epithelial cells (IEC) isolated from mice subjected to 17 days of treatment with vancomycin (V), neomycin (N), metronidazol (M), and amphotericin-B (Amf-B) every 12 hours by gavage with ampicillin (A) ad libitum in drinking water (VNMAmf-B+A) compared to mice gavaged with water only (Sham) and from colonic IEC of untreated germ-free mice (GF) and their conventional controls (Conv). Target gene listed on top of each panel and plotted in relation to the house-keeping gene Hprt. Each symbol represents one mouse, horizontal bars represent medians. Mann-Whitney analyses between VNMAmf-B+A and Sham, p<0.05 for all genes.
Similar articles
- Toll-like receptor 2 is critical for induction of Reg3 beta expression and intestinal clearance of Yersinia pseudotuberculosis.
Dessein R, Gironella M, Vignal C, Peyrin-Biroulet L, Sokol H, Secher T, Lacas-Gervais S, Gratadoux JJ, Lafont F, Dagorn JC, Ryffel B, Akira S, Langella P, Nùñez G, Sirard JC, Iovanna J, Simonet M, Chamaillard M. Dessein R, et al. Gut. 2009 Jun;58(6):771-6. doi: 10.1136/gut.2008.168443. Epub 2009 Jan 27. Gut. 2009. PMID: 19174417 - Modulation of Gut Microbiota Composition by Serotonin Signaling Influences Intestinal Immune Response and Susceptibility to Colitis.
Kwon YH, Wang H, Denou E, Ghia JE, Rossi L, Fontes ME, Bernier SP, Shajib MS, Banskota S, Collins SM, Surette MG, Khan WI. Kwon YH, et al. Cell Mol Gastroenterol Hepatol. 2019;7(4):709-728. doi: 10.1016/j.jcmgh.2019.01.004. Epub 2019 Feb 1. Cell Mol Gastroenterol Hepatol. 2019. PMID: 30716420 Free PMC article. - Comparison of different modes of antibiotic delivery on gut microbiota depletion efficiency and body composition in mouse.
Tirelle P, Breton J, Riou G, Déchelotte P, Coëffier M, Ribet D. Tirelle P, et al. BMC Microbiol. 2020 Nov 11;20(1):340. doi: 10.1186/s12866-020-02018-9. BMC Microbiol. 2020. PMID: 33176677 Free PMC article. - Intestinal MicrobiOMICS to define health and disease in human and mice.
Serino M, Chabo C, Burcelin R. Serino M, et al. Curr Pharm Biotechnol. 2012 Apr;13(5):746-58. doi: 10.2174/138920112799857567. Curr Pharm Biotechnol. 2012. PMID: 22122483 Review.
Cited by
- Intestinal stem cell niche: An upcoming area of immense importance in gastrointestinal disorders.
Mehra L, Bhowmik S, Makharia GK, Das P. Mehra L, et al. Indian J Gastroenterol. 2024 Nov 8. doi: 10.1007/s12664-024-01699-8. Online ahead of print. Indian J Gastroenterol. 2024. PMID: 39514159 Review. - A Ketogenic Diet Affects Gut Microbiota by Regulating Gut Microbiota and Promoting Hippocampal TRHR Expression to Combat Seizures.
Xiong W, Lin X, Lin X, Wu L, Lin W. Xiong W, et al. J Mol Neurosci. 2024 Nov 3;74(4):104. doi: 10.1007/s12031-024-02245-z. J Mol Neurosci. 2024. PMID: 39489848 - Transfer of microbiota from lean donors in combination with prebiotics prevents excessive weight gain and improves gut-brain vagal signaling in obese rats.
Minaya DM, Kim JS, Kirkland R, Allen J, Cullinan S, Maclang N, de Lartigue G, de La Serre C. Minaya DM, et al. Gut Microbes. 2024 Jan-Dec;16(1):2421581. doi: 10.1080/19490976.2024.2421581. Epub 2024 Nov 1. Gut Microbes. 2024. PMID: 39485288 Free PMC article. - MICROBIOME: The trials and errors of developing an experimental model to study the impact of maternal gut microbiome disruption on perinatal asphyxia.
Ionescu MI, Maria Catrina A, Dogaru IA, Catalina Barbalata D, Ciotei C, Haidoiu C, Suhaianu V, Gradisteanu Pircalabioru G, O'Mahony SM, Zagrean AM. Ionescu MI, et al. Reprod Fertil. 2024 Nov 6;5(4):e240050. doi: 10.1530/RAF-24-0050. Print 2024 Oct 1. Reprod Fertil. 2024. PMID: 39374132 Free PMC article. - Intestinal Tuft Cells Are Enriched With Protocadherins.
Stubler R, Dooley SA, Edens R, Nicholson MR, Engevik AC. Stubler R, et al. J Histochem Cytochem. 2024 Oct;72(10):611-622. doi: 10.1369/00221554241287267. Epub 2024 Oct 3. J Histochem Cytochem. 2024. PMID: 39360911
References
- Noverr MC, Huffnagle GB. Does the microbiota regulate immune responses outside the gut? Trends Microbiol. 2004;12:562–568. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources