Variation in tropical reef symbiont metagenomes defined by secondary metabolism - PubMed (original) (raw)
Variation in tropical reef symbiont metagenomes defined by secondary metabolism
Mohamed S Donia et al. PLoS One. 2011.
Abstract
The complex evolution of secondary metabolism is important in biology, drug development, and synthetic biology. To examine this problem at a fine scale, we compared the genomes and chemistry of 24 strains of uncultivated cyanobacteria, Prochloron didemni, that live symbiotically with tropical ascidians and that produce natural products isolated from the animals. Although several animal species were obtained along a >5500 km transect of the Pacific Ocean, P. didemni strains are >97% identical across much of their genomes, with only a few exceptions concentrated in secondary metabolism. Secondary metabolic gene clusters were sporadically present or absent in identical genomic locations with no consistent pattern of co-occurrence. Discrete mutations were observed, leading to new chemicals that we isolated from animals. Functional cassettes encoding diverse chemicals are exchanged among a single population of symbiotic P. didemni that spans the tropical Pacific, providing the host animals with a varying arsenal of secondary metabolites.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
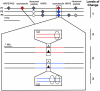
Figure 1. Secondary metabolic pathways from three Prochloron metagenomes.
Center: Prochloron cells (top) from the ascidian L. patella (bottom). Surrounding are ten pathway types identified, comprising 15 distinct biosynthetic gene clusters in the metagenomes of P1, P2 and P3. Compounds that have been chemically isolated are indicated by an asterisk; the remainder are genome-based predictions.
Figure 2. Variations in secondary metabolic gene clusters constitute the major difference between the P. didemni genomes.
a) The Artemis Comparison Tool (ACT) was used to visualize pair-wise comparisons between contig ASM-2318 from P1 and corresponding contigs from P2 and P3 . Violet color indicates ≥ 96% nucleotide identity while light blue color indicates 90–96% nucleotide identity. Two of the three differing regions marked by black boxes differ solely due to natural product gene clusters. This pattern is reproduced throughout the 3 metagenomes. b) and c) Dot Plot representations of NUCmer comparisons showing synteny between the same genetic regions as in A .
Figure 3. Schematic representation of the different levels of changes in functional cassettes in P. didemni genomes.
Three levels can be observed: 1. Presence or absence of a gene cluster in the genome. Homologous gene clusters are >99% identical between strains; 2. Swaps of function (heterocyclization to prenylation) within otherwise >99% identical gene clusters; 3. Functional point mutations in gene clusters (for example in product-coding sequences of ribosomal peptides).
Figure 4. Sporadic distribution of ascidian-associated secondary metabolic genes across the tropical Pacific.
Twenty-four samples collected over a 5,500 km transect in the tropical Pacific were screened for presence/absence of genes for five biosynthetic pathways. Each circle represents an individual ascidian sample. The shape inside the circle indicates the species of the _Prochloron_-harboring ascidian, as shown in the key above. Colors within the circle represent the presence of individual biosynthetic pathways, which are >99% identical between samples, while black indicates the absence of a pathway from the sample. S represents samples for which whole genome sequence was obtained.
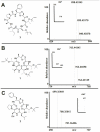
Figure 5. New prenylated cyanobactins discovered by comparative analysis.
A control known compound, trunkamide (3, panel a), was subjected to high-resolution mass spectrometry analysis. Using tandem mass spectrometry, both one and two isoprene side chains (C5H8, m/z = 68.0626) were eliminated from this molecule. In comparison, two new compounds, mollamide D (1) and mollamide E (2) from sample 03-002, also exhibited the same pattern (panels b and c). The high-resolution results corresponded both to the predicted masses of the compounds and to the predicted loss of isoprene.
Similar articles
- Complex microbiome underlying secondary and primary metabolism in the tunicate-Prochloron symbiosis.
Donia MS, Fricke WF, Partensky F, Cox J, Elshahawi SI, White JR, Phillippy AM, Schatz MC, Piel J, Haygood MG, Ravel J, Schmidt EW. Donia MS, et al. Proc Natl Acad Sci U S A. 2011 Dec 20;108(51):E1423-32. doi: 10.1073/pnas.1111712108. Epub 2011 Nov 28. Proc Natl Acad Sci U S A. 2011. PMID: 22123943 Free PMC article. - Natural combinatorial peptide libraries in cyanobacterial symbionts of marine ascidians.
Donia MS, Hathaway BJ, Sudek S, Haygood MG, Rosovitz MJ, Ravel J, Schmidt EW. Donia MS, et al. Nat Chem Biol. 2006 Dec;2(12):729-35. doi: 10.1038/nchembio829. Epub 2006 Nov 5. Nat Chem Biol. 2006. PMID: 17086177 - Excitation energy relaxation in a symbiotic cyanobacterium, Prochloron didemni, occurring in coral-reef ascidians, and in a free-living cyanobacterium, Prochlorothrix hollandica.
Hamada F, Yokono M, Hirose E, Murakami A, Akimoto S. Hamada F, et al. Biochim Biophys Acta. 2012 Nov;1817(11):1992-7. doi: 10.1016/j.bbabio.2012.06.008. Epub 2012 Jun 21. Biochim Biophys Acta. 2012. PMID: 22728755 - Population genetics of reef coral endosymbionts (Symbiodinium, Dinophyceae).
Thornhill DJ, Howells EJ, Wham DC, Steury TD, Santos SR. Thornhill DJ, et al. Mol Ecol. 2017 May;26(10):2640-2659. doi: 10.1111/mec.14055. Epub 2017 Mar 8. Mol Ecol. 2017. PMID: 28188662 Review. - Genome Evolution of Coral Reef Symbionts as Intracellular Residents.
González-Pech RA, Bhattacharya D, Ragan MA, Chan CX. González-Pech RA, et al. Trends Ecol Evol. 2019 Sep;34(9):799-806. doi: 10.1016/j.tree.2019.04.010. Epub 2019 May 10. Trends Ecol Evol. 2019. PMID: 31084944 Review.
Cited by
- Mechanism of Action of Ribosomally Synthesized and Post-Translationally Modified Peptides.
Ongpipattanakul C, Desormeaux EK, DiCaprio A, van der Donk WA, Mitchell DA, Nair SK. Ongpipattanakul C, et al. Chem Rev. 2022 Sep 28;122(18):14722-14814. doi: 10.1021/acs.chemrev.2c00210. Epub 2022 Sep 1. Chem Rev. 2022. PMID: 36049139 Free PMC article. Review. - Antiviral Peptides (AVPs) of Marine Origin as Propitious Therapeutic Drug Candidates for the Treatment of Human Viruses.
Sukmarini L. Sukmarini L. Molecules. 2022 Apr 19;27(9):2619. doi: 10.3390/molecules27092619. Molecules. 2022. PMID: 35565968 Free PMC article. Review. - Possible Functional Roles of Patellamides in the Ascidian-Prochloron Symbiosis.
Baur P, Kühl M, Comba P, Behrendt L. Baur P, et al. Mar Drugs. 2022 Feb 2;20(2):119. doi: 10.3390/md20020119. Mar Drugs. 2022. PMID: 35200648 Free PMC article. Review. - Thonningia sanguinea Extract: Antioxidant and Cytotoxic Activities Supported by Chemical Composition and Molecular Docking Simulations.
Abdelhameed RFA, Elhady SS, Sirwi A, Samir H, Ibrahim EA, Thomford AK, El Gindy A, Hadad GM, Badr JM, Nafie MS. Abdelhameed RFA, et al. Plants (Basel). 2021 Oct 11;10(10):2156. doi: 10.3390/plants10102156. Plants (Basel). 2021. PMID: 34685963 Free PMC article. - Assessing Biosynthetic Gene Cluster Diversity of Specialized Metabolites in the Conserved Gut Symbionts of Herbivorous Turtle Ants.
Chanson A, Moreau CS, Duplais C. Chanson A, et al. Front Microbiol. 2021 Jun 29;12:678100. doi: 10.3389/fmicb.2021.678100. eCollection 2021. Front Microbiol. 2021. PMID: 34267736 Free PMC article.
References
- Mayer AM, Glaser KB, Cuevas C, Jacobs RS, Kem W, et al. The odyssey of marine pharmaceuticals: a current pipeline perspective. Trends Pharmacol Sci. 2010;31:255–265. - PubMed
- Molinski TF, Dalisay DS, Lievens SL, Saludes JP. Drug development from marine natural products. Nat Rev Drug Discov. 2009;8:69–85. - PubMed
- Paul VJ, Ritson-Williams R. Marine chemical ecology. Nat Prod Rep. 2008;25:662–695. - PubMed
- Schmidt EW, Obraztsova AY, Davidson SK, Faulkner DJ, Haygood MG. Identification of the antifungal peptide-containing symbiont of the marine sponge Theonella swinhoei as a novel delta-proteobacterium, “Candidatus Entotheonella palauensis”. Marine Biology. 2000;136:969–977.
- Sudek S, Lopanik NB, Waggoner LE, Hildebrand M, Anderson C, et al. Identification of the putative bryostatin polyketide synthase gene cluster from “Candidatus Endobugula sertula”, the uncultivated microbial symbiont of the marine bryozoan Bugula neritina. J Nat Prod. 2007;70:67–74. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources