Low-coverage sequencing: implications for design of complex trait association studies - PubMed (original) (raw)
Low-coverage sequencing: implications for design of complex trait association studies
Yun Li et al. Genome Res. 2011 Jun.
Abstract
New sequencing technologies allow genomic variation to be surveyed in much greater detail than previously possible. While detailed analysis of a single individual typically requires deep sequencing, when many individuals are sequenced it is possible to combine shallow sequence data across individuals to generate accurate calls in shared stretches of chromosome. Here, we show that, as progressively larger numbers of individuals are sequenced, increasingly accurate genotype calls can be generated for a given sequence depth. We evaluate the implications of low-coverage sequencing for complex trait association studies. We systematically compare study designs based on genotyping of tagSNPs, sequencing of many individuals at depths ranging between 2× and 30×, and imputation of variants discovered by sequencing a subset of individuals into the remainder of the sample. We show that sequencing many individuals at low depth is an attractive strategy for studies of complex trait genetics. For example, for disease-associated variants with frequency >0.2%, sequencing 3000 individuals at 4× depth provides similar power to deep sequencing of >2000 individuals at 30× depth but requires only ~20% of the sequencing effort. We also show low-coverage sequencing can be used to build a reference panel that can drive imputation into additional samples to increase power further. We provide guidance for investigators wishing to combine results from sequenced, genotyped, and imputed samples.
Figures
Figure 1.
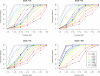
SNP discovery (%) by MAF, sequencing depth, and sequencing sample size. We simulated 30–2000 individuals sequenced at depths 2×, 4×, 6×, and 30×. We plotted the % of SNPs discovered by population MAF category (<0.1% to >5%), where population MAF is for the 45,000 simulated chromosomes. The dotted lines show the % of SNPs in each population MAF category that is polymorphic among the sequenced individuals.
Figure 2.
Genotype calling quality by MAF, sequencing depth, and sequencing sample size. We simulated 30–2000 individuals sequenced at depths 2×, 4×, and 6×. We compared genotype calls at detected SNPs with the simulated truth to obtain two measures of genotype calling quality, genotypic concordance and dosage r2, for each called SNP. We plot these two measures (left panel: genotypic concordance; right panel: dosage r2) by population MAF category (<0.1% to >5%), where population MAF is for the 45,000 simulated chromosomes.
Figure 3.
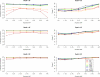
Genotype concordance and fraction of genotypes by non-ancestral allele counts (60 individuals sequenced at 4×). Genotype concordance (_y_-axis on left, dots) and fraction of genotypes (_y_-axis on right, bars) for simulated data, broken down by genotype category (homozygotes for the ancestral allele [HomRef], heterozygotes [Het], and homozygotes for the non-ancestral allele [HomAlt]) are plotted as a function of non-ancestral allele count among 60 individuals sequenced at 4× (_x_-axis).
Figure 4.
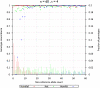
Genotype calling pipeline for the 1000 Genomes Pilot 1 Project. The pipeline we have developed to call genotypes for individuals sequenced at an average depth of ∼4–5× by the 1000 Genomes Pilot 1 Project.
Figure 5.
SNP detection power by minor allele count. For both simulated CEU and real data sets from the 1000 Genomes Project, SNPs were detected through a joint analysis of 59 or 60 individuals. Power of SNP detection was evaluated using a subset of 43 individuals.
Figure 6.
Genotype calling quality: Simulated versus the 1000 Genomes Pilot 1. Genotype calling quality is gauged by two measures—genotypic concordance and dosage r2—by comparing with true genotypes in simulated data and with experimental genotypes in real data from the 1000 Genomes Low-coverage Pilot Project. For both the real and simulated data, 60 individuals were sequenced at an average depth of 4×. For the 1000 Genomes Pilot 1 data, genotype calling was performed using sequencing data alone without HapMap 3 genotypes.
Figure 7.
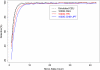
Power of association mapping by sequencing depth and number of individuals sequenced. We simulated 1500 cases and 1500 controls, assuming a single causal variant with causal allele frequency 0.5%, 1%, or 3%. We sequenced all 3000 individuals or a random subset of 400, 1000, or 2000 individuals (equal number of cases and controls) at depths ranging from 2×–30×. Power was estimated using an empirical threshold determined from 500 null sets to ensure familywise type-I error of 5%.
Similar articles
- Whole-genome characterization in pedigreed non-human primates using genotyping-by-sequencing (GBS) and imputation.
Bimber BN, Raboin MJ, Letaw J, Nevonen KA, Spindel JE, McCouch SR, Cervera-Juanes R, Spindel E, Carbone L, Ferguson B, Vinson A. Bimber BN, et al. BMC Genomics. 2016 Aug 24;17(1):676. doi: 10.1186/s12864-016-2966-x. BMC Genomics. 2016. PMID: 27558348 Free PMC article. - Comparing low-pass sequencing and genotyping for trait mapping in pharmacogenetics.
Wasik K, Berisa T, Pickrell JK, Li JH, Fraser DJ, King K, Cox C. Wasik K, et al. BMC Genomics. 2021 Mar 20;22(1):197. doi: 10.1186/s12864-021-07508-2. BMC Genomics. 2021. PMID: 33743587 Free PMC article. - Low-depth genotyping-by-sequencing (GBS) in a bovine population: strategies to maximize the selection of high quality genotypes and the accuracy of imputation.
Brouard JS, Boyle B, Ibeagha-Awemu EM, Bissonnette N. Brouard JS, et al. BMC Genet. 2017 Apr 5;18(1):32. doi: 10.1186/s12863-017-0501-y. BMC Genet. 2017. PMID: 28381212 Free PMC article. - Accurate Imputation of Untyped Variants from Deep Sequencing Data.
Torkamaneh D, Belzile F. Torkamaneh D, et al. Methods Mol Biol. 2021;2243:271-281. doi: 10.1007/978-1-0716-1103-6_13. Methods Mol Biol. 2021. PMID: 33606262 Review. - Genotype Imputation from Large Reference Panels.
Das S, Abecasis GR, Browning BL. Das S, et al. Annu Rev Genomics Hum Genet. 2018 Aug 31;19:73-96. doi: 10.1146/annurev-genom-083117-021602. Epub 2018 May 23. Annu Rev Genomics Hum Genet. 2018. PMID: 29799802 Review.
Cited by
- Genetic Risk for Alcohol Use Disorder in Relation to Individual Symptom Criteria: Do Polygenic Indices Provide Unique Information for Understanding Severity and Heterogeneity?
Kim Y, Lane SP, Miller AP, Wilhelmsen KC, Gizer IR. Kim Y, et al. medRxiv [Preprint]. 2024 Sep 23:2024.09.20.24313762. doi: 10.1101/2024.09.20.24313762. medRxiv. 2024. PMID: 39399010 Free PMC article. Preprint. - Modeling biases from low-pass genome sequencing to enable accurate population genetic inferences.
Fonseca EM, Tran LN, Mendoza H, Gutenkunst RN. Fonseca EM, et al. bioRxiv [Preprint]. 2024 Jul 23:2024.07.19.604366. doi: 10.1101/2024.07.19.604366. bioRxiv. 2024. PMID: 39091836 Free PMC article. Preprint. - Novel liquid biopsy CNV biomarkers in malignant melanoma.
Lukacova E, Hanzlikova Z, Podlesnyi P, Sedlackova T, Szemes T, Grendar M, Samec M, Hurtova T, Malicherova B, Leskova K, Budis J, Burjanivova T. Lukacova E, et al. Sci Rep. 2024 Jul 9;14(1):15786. doi: 10.1038/s41598-024-65928-y. Sci Rep. 2024. PMID: 38982214 Free PMC article. - Using blood transcriptome analysis for Alzheimer's disease diagnosis and patient stratification.
Zhong H, Zhou X, Uhm H, Jiang Y, Cao H, Chen Y, Mak TTW, Lo RMN, Wong BWY, Cheng EYL, Mok KY, Chan ALT, Kwok TCY, Mok VCT, Ip FCF, Hardy J, Fu AKY, Ip NY. Zhong H, et al. Alzheimers Dement. 2024 Apr;20(4):2469-2484. doi: 10.1002/alz.13691. Epub 2024 Feb 7. Alzheimers Dement. 2024. PMID: 38323937 Free PMC article. - The Born in Guangzhou Cohort Study enables generational genetic discoveries.
Huang S, Liu S, Huang M, He JR, Wang C, Wang T, Feng X, Kuang Y, Lu J, Gu Y, Xia X, Lin S; Born in Guangzhou Cohort Study (BIGCS) Group; Zhou W, Fu Q, Xia H, Qiu X. Huang S, et al. Nature. 2024 Feb;626(7999):565-573. doi: 10.1038/s41586-023-06988-4. Epub 2024 Jan 31. Nature. 2024. PMID: 38297123
References
- Baum LE 1972. An inequality and associated maximization technique in statistical estimation for probabilistic functions of Markov processes. Inequalities 3: 1–8
- Bentley DR 2006. Whole-genome re-sequencing. Curr Opin Genet Dev 16: 545–552 - PubMed
Publication types
MeSH terms
Grants and funding
- R01 CA082659/CA/NCI NIH HHS/United States
- R01 HG006292/HG/NHGRI NIH HHS/United States
- R01 HG000376/HG/NHGRI NIH HHS/United States
- R56 HG000376/HG/NHGRI NIH HHS/United States
- 3-R01-CA082659-11S1/CA/NCI NIH HHS/United States
- R01 CA082659-13/CA/NCI NIH HHS/United States
- HG000376/HG/NHGRI NIH HHS/United States
LinkOut - more resources
Full Text Sources