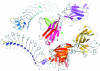Evaluating the solution from MrBUMP and BALBES - PubMed (original) (raw)
Evaluating the solution from MrBUMP and BALBES
Ronan M Keegan et al. Acta Crystallogr D Biol Crystallogr. 2011 Apr.
Abstract
Molecular replacement is one of the key methods used to solve the problem of determining the phases of structure factors in protein structure solution from X-ray image diffraction data. Its success rate has been steadily improving with the development of improved software methods and the increasing number of structures available in the PDB for use as search models. Despite this, in cases where there is low sequence identity between the target-structure sequence and that of its set of possible homologues it can be a difficult and time-consuming chore to isolate and prepare the best search model for molecular replacement. MrBUMP and BALBES are two recent developments from CCP4 that have been designed to automate and speed up the process of determining and preparing the best search models and putting them through molecular replacement. Their intention is to provide the user with a broad set of results using many search models and to highlight the best of these for further processing. An overview of both programs is presented along with a description of how best to use them, citing case studies and the results of large-scale testing of the software.
Figures
Figure 1
The types and hierarchy of the search models presented in BALBES jobs.
Figure 2
A flow chart of the structural solution process in BALBES.
Figure 3
An example shows how BALBES combines models from different PDB files to obtain a complex solution.
Figure 4
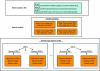
(a) The top-level directory structure for a MrBUMP job. All job information is stored under a folder ‘search_JobID’, where JobID is the job identifier. The ‘data’ directory contains all of the processing data for each of the template search models. (b) The breakdown of the ‘data’ directory. There are potentially four model types created for each template search model. Each model-type directory contains the molecular-replacement and refinement files for that model.
Figure 5
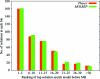
This histogram shows a breakdown of where the top-scoring solution in each of the MrBUMP jobs ranked in terms of its initial ranking before being put through molecular replacement. This ranking is based on the score function for the search models. As can be seen, it often happens that the top-scoring solution lies outside the first five top-scoring search models.
Figure 6
A plot of the model types for the best-scoring search models after molecular replacement and refinement for each of the MrBUMP jobs in the test set. We only show results where a solution was deemed to be ‘marginal’ or better.
Figure 7
Example
2wfb
: crystal structure of the apo form of the orange protein (Apo-Orp) from Desulfovibrio gigas (S. Najmudin, C. Bonifacio, A. G. Duarte, I. Moura, J. G. Moura & M. G. Romao, unpublished work). This is an ensemble example where the constituent search models (
1rdu
_ A,
1eo1
_ A,
1o13
_ A,
1t3v
_ A) in the ensemble failed to yield an acceptable solution on their own using Phaser to perform the MR. The ensemble itself produced a ‘good’ solution. Providing Phaser with the ensemble of aligned search models allowed it to downweight the signal from parts of the models which are not conserved across all of the structures, i.e. flexible loops and terminal residues. The figure was prepared using CCP_4_mg (Potterton et al., 2004 ▶).
Figure 8
Example
2zzt
: crystal structure of the cytosolic domain of the cation-diffusion facilitator family protein. The smaller domain in
2qfi
(zinc transporter YiiP; green) matches the structure of
2zzt
(pink) very closely in its secondary-structure conformation. The figure was prepared using CCP_4_mg (Potterton et al., 2004 ▶).
Similar articles
- Ensembles generated from crystal structures of single distant homologues solve challenging molecular-replacement cases in AMPLE.
Rigden DJ, Thomas JMH, Simkovic F, Simpkin A, Winn MD, Mayans O, Keegan RM. Rigden DJ, et al. Acta Crystallogr D Struct Biol. 2018 Mar 1;74(Pt 3):183-193. doi: 10.1107/S2059798318002310. Epub 2018 Mar 2. Acta Crystallogr D Struct Biol. 2018. PMID: 29533226 Free PMC article. - Recent developments in MrBUMP: better search-model preparation, graphical interaction with search models, and solution improvement and assessment.
Keegan RM, McNicholas SJ, Thomas JMH, Simpkin AJ, Simkovic F, Uski V, Ballard CC, Winn MD, Wilson KS, Rigden DJ. Keegan RM, et al. Acta Crystallogr D Struct Biol. 2018 Mar 1;74(Pt 3):167-182. doi: 10.1107/S2059798318003455. Epub 2018 Mar 6. Acta Crystallogr D Struct Biol. 2018. PMID: 29533225 Free PMC article. - SIMBAD: a sequence-independent molecular-replacement pipeline.
Simpkin AJ, Simkovic F, Thomas JMH, Savko M, Lebedev A, Uski V, Ballard C, Wojdyr M, Wu R, Sanishvili R, Xu Y, Lisa MN, Buschiazzo A, Shepard W, Rigden DJ, Keegan RM. Simpkin AJ, et al. Acta Crystallogr D Struct Biol. 2018 Jul 1;74(Pt 7):595-605. doi: 10.1107/S2059798318005752. Epub 2018 Jun 8. Acta Crystallogr D Struct Biol. 2018. PMID: 29968670 Free PMC article. - Ongoing developments in CCP4 for high-throughput structure determination.
Winn MD, Ashton AW, Briggs PJ, Ballard CC, Patel P. Winn MD, et al. Acta Crystallogr D Biol Crystallogr. 2002 Nov;58(Pt 11):1929-36. doi: 10.1107/s0907444902016116. Epub 2002 Oct 21. Acta Crystallogr D Biol Crystallogr. 2002. PMID: 12393924 Review. - Molecular replacement: tricks and treats.
Abergel C. Abergel C. Acta Crystallogr D Biol Crystallogr. 2013 Nov;69(Pt 11):2167-73. doi: 10.1107/S0907444913015291. Epub 2013 Oct 12. Acta Crystallogr D Biol Crystallogr. 2013. PMID: 24189227 Free PMC article. Review.
Cited by
- Structural, mechanistic, and physiological insights into phospholipase A-mediated membrane phospholipid degradation in Pseudomonas aeruginosa.
Bleffert F, Granzin J, Caliskan M, Schott-Verdugo SN, Siebers M, Thiele B, Rahme L, Felgner S, Dörmann P, Gohlke H, Batra-Safferling R, Jaeger KE, Kovacic F. Bleffert F, et al. Elife. 2022 May 10;11:e72824. doi: 10.7554/eLife.72824. Elife. 2022. PMID: 35536643 Free PMC article. - Molecular replacement then and now.
Scapin G. Scapin G. Acta Crystallogr D Biol Crystallogr. 2013 Nov;69(Pt 11):2266-75. doi: 10.1107/S0907444913011426. Epub 2013 Oct 18. Acta Crystallogr D Biol Crystallogr. 2013. PMID: 24189239 Free PMC article. Review. - Cloning, expression, purification, crystallization and preliminary X-ray diffraction analysis of a ferredoxin/flavodoxin-NADP(H) oxidoreductase (Bc0385) from Bacillus cereus.
Skråmo S, Hersleth HP, Hammerstad M, Andersson KK, Røhr ÅK. Skråmo S, et al. Acta Crystallogr F Struct Biol Commun. 2014 Jun;70(Pt 6):777-80. doi: 10.1107/S2053230X14008334. Epub 2014 May 10. Acta Crystallogr F Struct Biol Commun. 2014. PMID: 24915092 Free PMC article. - Ensembles generated from crystal structures of single distant homologues solve challenging molecular-replacement cases in AMPLE.
Rigden DJ, Thomas JMH, Simkovic F, Simpkin A, Winn MD, Mayans O, Keegan RM. Rigden DJ, et al. Acta Crystallogr D Struct Biol. 2018 Mar 1;74(Pt 3):183-193. doi: 10.1107/S2059798318002310. Epub 2018 Mar 2. Acta Crystallogr D Struct Biol. 2018. PMID: 29533226 Free PMC article. - ContaMiner and ContaBase: a webserver and database for early identification of unwantedly crystallized protein contaminants.
Hungler A, Momin A, Diederichs K, Arold ST. Hungler A, et al. J Appl Crystallogr. 2016 Nov 2;49(Pt 6):2252-2258. doi: 10.1107/S1600576716014965. eCollection 2016 Dec 1. J Appl Crystallogr. 2016. PMID: 27980519 Free PMC article.
References
- Allan, R. J., Nave, C., Keegan, R. M., Meredith, D. J., Winn, M. D., Winter, G., Dolomanov, O., Launer, L., Young, P. & Berry, I. (2005). In Proceedings of the UK e-Science All Hands Meeting. http://www.allhands.org.uk/2005/proceedings/.
- Cheung, T. Y., Fairchild, M. J., Zarivach, R., Tanentzapf, G. & Van Petegem, F. (2009). J. Mol. Biol. 387, 787–793. - PubMed
- Coffey, A. & Ross, R. P. (2002). Antonie Van Leeuwenhoek, 82, 303–321. - PubMed
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- WT_/Wellcome Trust/United Kingdom
- BB/F020228/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/F020805/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
LinkOut - more resources
Full Text Sources