Presenting your structures: the CCP4mg molecular-graphics software - PubMed (original) (raw)
Presenting your structures: the CCP4mg molecular-graphics software
S McNicholas et al. Acta Crystallogr D Biol Crystallogr. 2011 Apr.
Abstract
CCP4mg is a molecular-graphics program that is designed to give rapid access to both straightforward and complex static and dynamic representations of macromolecular structures. It has recently been updated with a new interface that provides more sophisticated atom-selection options and a wizard to facilitate the generation of complex scenes. These scenes may contain a mixture of coordinate-derived and abstract graphical objects, including text objects, arbitrary vectors, geometric objects and imported images, which can enhance a picture and eliminate the need for subsequent editing. Scene descriptions can be saved to file and transferred to other molecules. Here, the substantially enhanced version 2 of the program, with a new underlying GUI toolkit, is described. A built-in rendering module produces publication-quality images.
Figures
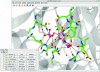
Figure 1
The CCP_4_mg main window and display table. Scene automatically created with Picture wizard style ‘ligand binding site: site and broken ribbons’ for PDB entry
1snf
(Chan et al., 2004 ▶).
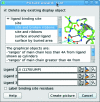
Figure 2
The Picture wizard. The selected style automatically created the scene shown in Fig. 1 ▶.
Figure 3
The electron-density styles possible in CCP_4_mg: (a) chickenwire, (b) chickenwire cylinders, (c) dots, (d) solid and (e) solid with transparency.
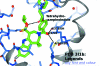
Figure 4
The text-display capabilities of CCP_4_mg. Three atoms are labelled with the default format: chain/residue number/atom name. The ligand has an annotation attached. All these rotate as the view is changed. In contrast, the ‘Legend’ is in a static user-defined screen position.
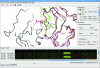
Figure 5
Structure and sequence alignment. Chain A of PDB entry
1eu5
(brown; González et al., 2001 ▶) superimposed on chain B of
2xce
(pink; García-Nafría et al., 2010 ▶) using SSM. The nucleotide analogue dUpNpp bound to
2xce
is drawn as cylinders. The sequences, aligned using MUSCLE, are displayed in the ‘Sequence viewer’ window coloured by conservation. The residues highlighted in the sequence viewer appear as stick representations in the graphics window. This figure also demonstrates the new single-window appearance of CCP_4_mg, with the Sequence viewer, Display Table and Clip and Fog widgets docked in the main window.
Figure 6
Assembly prediction by PISA. (a) The original model (PDB entry
3hhz
; Green & Luo, 2009 ▶). (b) The dimer predicted by PISA.
Figure 7
The Movie editor dialogue. The above sequence rotates an expanded view of the molecule around the y axis for 2 s, takes 2 s to zoom into a close-up view of a ligand and then rocks for 2 s with the ligand at the screen centre.
Similar articles
- Coot: model-building tools for molecular graphics.
Emsley P, Cowtan K. Emsley P, et al. Acta Crystallogr D Biol Crystallogr. 2004 Dec;60(Pt 12 Pt 1):2126-32. doi: 10.1107/S0907444904019158. Epub 2004 Nov 26. Acta Crystallogr D Biol Crystallogr. 2004. PMID: 15572765 - Recent developments in MrBUMP: better search-model preparation, graphical interaction with search models, and solution improvement and assessment.
Keegan RM, McNicholas SJ, Thomas JMH, Simpkin AJ, Simkovic F, Uski V, Ballard CC, Winn MD, Wilson KS, Rigden DJ. Keegan RM, et al. Acta Crystallogr D Struct Biol. 2018 Mar 1;74(Pt 3):167-182. doi: 10.1107/S2059798318003455. Epub 2018 Mar 6. Acta Crystallogr D Struct Biol. 2018. PMID: 29533225 Free PMC article. - Evaluating the solution from MrBUMP and BALBES.
Keegan RM, Long F, Fazio VJ, Winn MD, Murshudov GN, Vagin AA. Keegan RM, et al. Acta Crystallogr D Biol Crystallogr. 2011 Apr;67(Pt 4):313-23. doi: 10.1107/S0907444911007530. Epub 2011 Mar 18. Acta Crystallogr D Biol Crystallogr. 2011. PMID: 21460449 Free PMC article. - A visual data flow environment for macromolecular crystallographic computing.
Wild DL, Tucker PA, Choe S. Wild DL, et al. J Mol Graph. 1995 Oct;13(5):291-8, 299-300. doi: 10.1016/0263-7855(95)00068-2. J Mol Graph. 1995. PMID: 8603058 - Boxes of Model Building and Visualization.
Turk D. Turk D. Methods Mol Biol. 2017;1607:491-548. doi: 10.1007/978-1-4939-7000-1_21. Methods Mol Biol. 2017. PMID: 28573587 Review.
Cited by
- Ultrasonic acoustic levitation for fast frame rate X-ray protein crystallography at room temperature.
Tsujino S, Tomizaki T. Tsujino S, et al. Sci Rep. 2016 May 6;6:25558. doi: 10.1038/srep25558. Sci Rep. 2016. PMID: 27150272 Free PMC article. - Crystal Structure of the Pseudomonas aeruginosa BEL-1 Extended-Spectrum β-Lactamase and Its Complexes with Moxalactam and Imipenem.
Pozzi C, De Luca F, Benvenuti M, Poirel L, Nordmann P, Rossolini GM, Mangani S, Docquier JD. Pozzi C, et al. Antimicrob Agents Chemother. 2016 Nov 21;60(12):7189-7199. doi: 10.1128/AAC.00936-16. Print 2016 Dec. Antimicrob Agents Chemother. 2016. PMID: 27671060 Free PMC article. - Crystallographic fragment-binding studies of the Mycobacterium tuberculosis trifunctional enzyme suggest binding pockets for the tails of the acyl-CoA substrates at its active sites and a potential substrate-channeling path between them.
Dalwani S, Metz A, Huschmann FU, Weiss MS, Wierenga RK, Venkatesan R. Dalwani S, et al. Acta Crystallogr D Struct Biol. 2024 Aug 1;80(Pt 8):605-619. doi: 10.1107/S2059798324006557. Epub 2024 Jul 16. Acta Crystallogr D Struct Biol. 2024. PMID: 39012716 Free PMC article. - Epitope Mapping of Exposed Tegument and Alimentary Tract Proteins Identifies Putative Antigenic Targets of the Attenuated Schistosome Vaccine.
Farias LP, Vance GM, Coulson PS, Vitoriano-Souza J, Neto APDS, Wangwiwatsin A, Neves LX, Castro-Borges W, McNicholas S, Wilson KS, Leite LCC, Wilson RA. Farias LP, et al. Front Immunol. 2021 Mar 3;11:624613. doi: 10.3389/fimmu.2020.624613. eCollection 2020. Front Immunol. 2021. PMID: 33763055 Free PMC article. - Structural and mechanistic characterization of bifunctional heparan sulfate N-deacetylase-N-sulfotransferase 1.
Mycroft-West CJ, Abdelkarim S, Duyvesteyn HME, Gandhi NS, Skidmore MA, Owens RJ, Wu L. Mycroft-West CJ, et al. Nat Commun. 2024 Feb 13;15(1):1326. doi: 10.1038/s41467-024-45419-4. Nat Commun. 2024. PMID: 38351061 Free PMC article.
References
- Abagyan, R., Lee, W. H., Raush, E., Budagyan, L., Totrov, M., Sundstrom, M. & Marsden, B. D. (2006). Trends Biochem. Sci. 31, 76–78. - PubMed
- Artymiuk, P. J., Rice, D. W., Mitchell, M. E. & Willett, P. (1989). J. Inform. Sci. 15, 287–298.
- Bernstein, H. J. (2000). Trends Biochem. Sci. 25, 453–455. - PubMed
- Carson, M. (1997). Methods Enzymol. 277, 493–505. - PubMed
- Chan, S. et al. (2004). J. Mol. Biol. 341, 503–517. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources