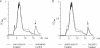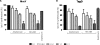Mechanism of action and limited cross-resistance of new lipopeptide MX-2401 - PubMed (original) (raw)
. 2011 Jun;55(6):2743-54.
doi: 10.1128/AAC.00170-11. Epub 2011 Apr 4.
T Schneider, M Elliott, W R P Scott, J Pan, C Anklin, H Yang, D Dugourd, A Müller, K Gries, S K Straus, H G Sahl, R E W Hancock
Affiliations
- PMID: 21464247
- PMCID: PMC3101398
- DOI: 10.1128/AAC.00170-11
Mechanism of action and limited cross-resistance of new lipopeptide MX-2401
E Rubinchik et al. Antimicrob Agents Chemother. 2011 Jun.
Abstract
MX-2401 is a semisynthetic calcium-dependent lipopeptide antibiotic (analogue of amphomycin) in preclinical development for the treatment of serious Gram-positive infections. In vitro and in vivo, MX-2401 demonstrates broad-spectrum bactericidal activity against Gram-positive organisms, including antibiotic-resistant strains. The objective of this study was to investigate the mechanism of action of MX-2401 and compare it with that of the lipopeptide daptomycin. The results indicated that although both daptomycin and MX-2401 are in the structural class of Ca²⁺-dependent lipopeptide antibiotics, the latter has a different mechanism of action. Specifically, MX-2401 inhibits peptidoglycan synthesis by binding to the substrate undecaprenylphosphate (C₅₅-P), the universal carbohydrate carrier involved in several biosynthetic pathways. This interaction resulted in inhibition, in a dose-dependent manner, of the biosynthesis of the cell wall precursors lipids I and II and the wall teichoic acid precursor lipid III, while daptomycin had no significant effect on these processes. MX-2401 induced very slow membrane depolarization that was observed only at high concentrations. Unlike daptomycin, membrane depolarization by MX-2401 did not correlate with its bactericidal activity and did not affect general membrane permeability. In contrast to daptomycin, MX-2401 had no effect on lipid flip-flop, calcein release, or membrane fusion with 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC)/1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-(1'-rac-glycerol) (sodium salt) (POPG) liposomes. MX-2401 adopts a more defined structure than daptomycin, presumably to facilitate interaction with C₅₅-P. Mutants resistant to MX-2401 demonstrated low cross-resistance to other antibiotics. Overall, these results provided strong evidence that the mode of action of MX-2401 is unique and different from that of any of the approved antibiotics, including daptomycin.
Figures
Fig. 1.
MX-2401 structure.
Fig. 2.
Intracellular accumulation of UDP-_N_-acetylmuramyl pentapeptide in untreated and vancomycin-treated (A) and amphomycin-treated and MX-2401-treated (B) S. simulans 22 cells. The experiment was performed with 10× MIC of the respective antibiotic compounds for 45 min. Subsequently, cells were extracted with boiling water, and the intracellular nucleotide pool was analyzed by reverse-phase HPLC. The identity of UDP-MurNAc-pentapeptide was confirmed by MALDI-TOF mass spectrometry, yielding a molecular mass of 1,148.72 Da for the indicated peaks (arrows).
Fig. 3.
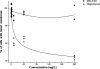
Impacts of amphomycin and MX-2401 on the overall in vitro lipid II synthesis by cytoplasmic membrane preparations of M. luteus. (A) The synthesis of lipid II in the presence of increasing concentrations of amphomycin and MX2401 was qualitatively analyzed using TLC and PMA staining. (B) Quantitative analysis was carried out using [14C]UDP-GlcNAc as described in Materials and Methods. Peptides were added at molar ratios of 0.25 to 2 with respect to the amount of the substrate C55-P. The reaction product of the untreated control was taken as 100%. The error bars indicate standard deviations.
Fig. 4.
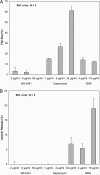
Impacts of amphomycin and MX-2401 on bactoprenol-phosphate-consuming reactions catalyzed by MraY (A) and TagO (B). The incorporation of [14C]UDP-MurNAc-pentapeptide into lipid I (A) and [14C]UDP-GlcNAc into lipid III (B) was analyzed in the presence of increasing peptide concentrations. Peptides were added in molar ratios of 0.25 to 2 with respect to the substrate C55-P. Daptomycin was added in 2-fold molar excess with respect to C55-P. Quantification was carried out using phosphorimaging. The error bars indicate standard deviations.
Fig. 5.
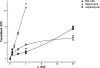
Dose-dependent membrane depolarization by MX-2401, daptomycin, and amphomycin. Cells were incubated with antibiotics for 60 min before depolarization was measured.
Fig. 6.
Membrane depolarization and cell death in S. epidermidis following incubation with sub- and supra-MIC concentrations of antibiotics in the presence or absence of surfactant. (A) Cells incubated with MX-2401 for 90 min. (B) Cells incubated with amphomycin for 90 min. (C) Cells incubated with daptomycin for 60 min.
Fig. 7.
Effects of MX-2401 and daptomycin on bacterial membrane permeability.
Fig. 8.
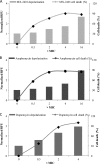
Ability of MX-2401 to interact with model membranes. (A) Dose-dependent lipid flip-flop of C6-NBD-PC asymmetrically labeled POPC/POPG liposomes treated with MX-2401, daptomycin, and GMS in the presence of 5 mM CaCl2. (B) Dose-dependent calcein release from POPC/POPG liposomes treated with MX-2401, daptomycin, and GMS in the presence of 5 mM CaCl2. The error bars indicate standard deviations.
Fig. 9.
Effect of MX-2401 and daptomycin on membrane fusion as determined by light scattering. Daptomycin (black bars) induces fusion in the presence of POPC, POPG, and Ca2+ as observed by the increase in vesicle size, whereas MX-2401 (white bars) does not, similarly to the control (gray bars; lipid only). The error bars represent the error associated with three repeats.
Fig. 10.
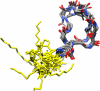
Representative structures of MX-2401 as obtained from the refinement simulations. The backbone trace on the right represents residues Asp1 → Pro11 (carbon, gray; nitrogen, blue; and oxygen, red), while the traces on the left (yellow) represent the acyl chain and linker connected to the N-terminal residue Asp1. The cyclic part is well defined (relative to daptomycin) and appears to adopt two major conformations. The image was created using UCSF CHIMERA (26).
Similar articles
- Antimicrobial properties of MX-2401, an expanded-spectrum lipopeptide active in the presence of lung surfactant.
Dugourd D, Yang H, Elliott M, Siu R, Clement JJ, Straus SK, Hancock RE, Rubinchik E. Dugourd D, et al. Antimicrob Agents Chemother. 2011 Aug;55(8):3720-8. doi: 10.1128/AAC.00322-11. Epub 2011 May 16. Antimicrob Agents Chemother. 2011. PMID: 21576435 Free PMC article. - Structure-Activity Relationships of Daptomycin Lipopeptides.
Karas JA, Carter GP, Howden BP, Turner AM, Paulin OKA, Swarbrick JD, Baker MA, Li J, Velkov T. Karas JA, et al. J Med Chem. 2020 Nov 25;63(22):13266-13290. doi: 10.1021/acs.jmedchem.0c00780. Epub 2020 Aug 6. J Med Chem. 2020. PMID: 32687352 Review. - The action mechanism of daptomycin.
Taylor SD, Palmer M. Taylor SD, et al. Bioorg Med Chem. 2016 Dec 15;24(24):6253-6268. doi: 10.1016/j.bmc.2016.05.052. Epub 2016 May 28. Bioorg Med Chem. 2016. PMID: 27288182 Review. - Daptomycin Leakage Is Selective.
Zhang J, Scoten K, Straus SK. Zhang J, et al. ACS Infect Dis. 2016 Oct 14;2(10):682-687. doi: 10.1021/acsinfecdis.6b00152. Epub 2016 Sep 28. ACS Infect Dis. 2016. PMID: 27669740 - Correlation of daptomycin resistance in a clinical Staphylococcus aureus strain with increased cell wall teichoic acid production and D-alanylation.
Bertsche U, Weidenmaier C, Kuehner D, Yang SJ, Baur S, Wanner S, Francois P, Schrenzel J, Yeaman MR, Bayer AS. Bertsche U, et al. Antimicrob Agents Chemother. 2011 Aug;55(8):3922-8. doi: 10.1128/AAC.01226-10. Epub 2011 May 23. Antimicrob Agents Chemother. 2011. PMID: 21606222 Free PMC article.
Cited by
- Bacillus subtilis uses the SigM signaling pathway to prioritize the use of its lipid carrier for cell wall synthesis.
Roney IJ, Rudner DZ. Roney IJ, et al. PLoS Biol. 2024 Apr 29;22(4):e3002589. doi: 10.1371/journal.pbio.3002589. eCollection 2024 Apr. PLoS Biol. 2024. PMID: 38683856 Free PMC article. - Synthesis and characterization of the 47-residue heterodimeric antimicrobial peptide distinctin, featuring directed disulfide bridge formation.
Mullen DG, Verardi R, Porcelli F, Scaloni A, Barany G, Veglia G. Mullen DG, et al. Biopolymers. 2012;98(5):479-84. doi: 10.1002/bip.22087. Biopolymers. 2012. PMID: 23203692 Free PMC article. - The Bacillus subtilis cell envelope stress-inducible ytpAB operon modulates membrane properties and contributes to bacitracin resistance.
Willdigg JR, Patel Y, Arquilevich BE, Subramanian C, Frank MW, Rock CO, Helmann JD. Willdigg JR, et al. J Bacteriol. 2024 Mar 21;206(3):e0001524. doi: 10.1128/jb.00015-24. Epub 2024 Feb 7. J Bacteriol. 2024. PMID: 38323910 Free PMC article. - Identification and in vitro analysis of the GatD/MurT enzyme-complex catalyzing lipid II amidation in Staphylococcus aureus.
Münch D, Roemer T, Lee SH, Engeser M, Sahl HG, Schneider T. Münch D, et al. PLoS Pathog. 2012 Jan;8(1):e1002509. doi: 10.1371/journal.ppat.1002509. Epub 2012 Jan 26. PLoS Pathog. 2012. PMID: 22291598 Free PMC article. - Wall teichoic acids of gram-positive bacteria.
Brown S, Santa Maria JP Jr, Walker S. Brown S, et al. Annu Rev Microbiol. 2013;67:313-36. doi: 10.1146/annurev-micro-092412-155620. Annu Rev Microbiol. 2013. PMID: 24024634 Free PMC article. Review.
References
- Baltz R. H., Miao V., Wrigley S. K. 2005. Natural products to drugs: daptomycin and related lipopeptide antibiotics. Nat. Prod. Rep. 22:717–741 - PubMed
- Berendsen H. J. C., Postma J. P. M., van Gunsteren W. F., Dinola A., Haak J. R. 1984. Molecular-dynamics with coupling to an external bath. J. Chem. Phys. 81:3684–3690
- Braunschweiler L., Ernst R. R. 1983. Coherence transfer by isotropic mixing: application to proton correlation spectroscopy. J. Magn. Reson. 53:521–528
- Brötz H., et al. 1998. Role of lipid-bound peptidoglycan precursors in the formation of pores by nisin, epidermin and other lantibiotics. Mol. Microbiol. 30:317–327 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous