Transcriptional regulation of pro-apoptotic protein kinase Cdelta: implications for oxidative stress-induced neuronal cell death - PubMed (original) (raw)
Transcriptional regulation of pro-apoptotic protein kinase Cdelta: implications for oxidative stress-induced neuronal cell death
Huajun Jin et al. J Biol Chem. 2011.
Abstract
We previously demonstrated that protein kinase Cδ (PKCδ; PKC delta) is an oxidative stress-sensitive kinase that plays a causal role in apoptotic cell death in neuronal cells. Although PKCδ activation has been extensively studied, relatively little is known about the molecular mechanisms controlling PKCδ expression. To characterize the regulation of PKCδ expression, we cloned an ∼2-kbp 5'-promoter segment of the mouse Prkcd gene. Deletion analysis indicated that the noncoding exon 1 region contained multiple Sp sites, including four GC boxes and one CACCC box, which directed the highest levels of transcription in neuronal cells. In addition, an upstream regulatory region containing adjacent repressive and anti-repressive elements with opposing regulatory activities was identified within the region -712 to -560. Detailed mutagenesis studies revealed that each Sp site made a positive contribution to PKCδ promoter expression. Overexpression of Sp family proteins markedly stimulated PKCδ promoter activity without any synergistic transactivating effect. Furthermore, experiments in Sp-deficient SL2 cells indicated long isoform Sp3 as the essential activator of PKCδ transcription. Importantly, both PKCδ promoter activity and endogenous PKCδ expression in NIE115 cells and primary striatal cultures were inhibited by mithramycin A. The results from chromatin immunoprecipitation and gel shift assays further confirmed the functional binding of Sp proteins to the PKCδ promoter. Additionally, we demonstrated that overexpression of p300 or CREB-binding protein increases the PKCδ promoter activity. This stimulatory effect requires intact Sp-binding sites and is independent of p300 histone acetyltransferase activity. Finally, modulation of Sp transcriptional activity or protein level profoundly altered the cell death induced by oxidative insult, demonstrating the functional significance of Sp-dependent PKCδ gene expression. Collectively, our findings may have implications for development of new translational strategies against oxidative damage.
Figures
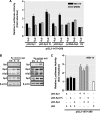
FIGURE 1.
Deletion analysis of PKCδ promoter activity in NIE115 and MN9D cells. A, schematic diagram of mouse PKCδ gene structure on chromosome 14. Exons are marked by boxes and are numbered below each box, and black and white regions within the boxes indicate the coding and noncoding exons, respectively. Arrow indicates the position of the translation start codon (ATG). B, schematic representation of PKCδ promoter deletion/luciferase reporter constructs. An extensive series of PKCδ promoter deletion derivatives was generated by PCR methods and inserted into the pGL3-Basic luciferase vector. The 5′- and 3′-positions of the constructs with respect to the transcription start site are depicted. C, each construct as shown in B was transiently transfected into NIE115 (black bar) and MN9D (white bar) cells. Cells were harvested 24 h after transfection, and luciferase activities were determined. The plasmid pcDNA3.1-βgal was included in each transfection to normalize the promoter activity with transfection efficiency. The activity of full-length promoter construct (pGL3−1694/+289) was arbitrarily set to 100, and the relative luciferase activity of the other constructs was calculated accordingly. The results represent the mean ± S.E. of three independent experiments performed in triplicate.
FIGURE 2.
Mapping of the identified repressive and anti-repressive elements within the PKCδ promoter and evidence for the PKCδ promoter-specific repressive element. A, schematic representation of PKCδ promoter 5′-deletion constructs used for the fine mapping study. The 5′- and 3′-positions of the constructs with respect to the transcription start site are depicted. B, each construct as depicted in A was transiently transfected into NIE115 (black bar) and MN9D (white bar) cells. Cells were harvested 24 h after transfection for assaying luciferase activities. The plasmid pcDNA3.1-βgal was cotransfected into cells for data normalization. The activity of pGL3−147/+289 was arbitrarily set to 100, and the relative luciferase activity of the other constructs is presented. The results represent the mean ± S.E. of three independent experiments performed in triplicate. C, isolated repressive element of the PKCδ promoter does not function as a locus-independent DNA element. The sequences around the identified repressive element (−660 to −561 of the PKCδ promoter) were directly fused to the 5′-end of the region between −147 to +289 of the PKCδ promoter and cloned into the pGL3-Basic luciferase vector to obtain pGL3−660/−561 plus −147/+289. NIE115 (black bar) and MN9D cells (white bar) were transfected with pGL3−147/+289 or pGL3−660/−561 plus −147/+289 for 24 h, and luciferase activity was determined. Schematic diagram of these constructs are shown at the right. The activity of pGL3−147/+289 was set to 100, and the relative luciferase activity of pGL3−660/−561 plus −147/+289 is presented. The results represent the mean ± S.E. of three independent experiments performed in triplicate. D, isolated repressive element of the PKCδ promoter does not act on a heterologous promoter (SV40). The sequences of the putative PKCδ repressive element (−660 to −561 of the PKCδ promoter) were cloned upstream of the SV40 promoter in pGL3−promoter vector to obtain pGL3−promoter-660/−561. NIE115 (black bar) and MN9D (white bar) cells were transfected with pGL3−promoter or pGL3−promoter-660/−561 for 24 h, and luciferase activity was determined. Schematic diagram of these constructs are shown at the right. The activity of pGL3−promoter was set to 100, and the relative luciferase activity of pGL3−promoter-660/−561 is given. The results represent the mean ± S.E. of three independent experiments performed in triplicate.
FIGURE 3.
Functional analysis of the PKCδ proximal promoter. A, sequence comparison of the mouse PKCδ promoter region between +2 to +289 with the corresponding regions of the rat and human PKCδ promoters. Sequences were aligned with the DiAlign TF program. Sequence differences are indicated, and gaps introduced to maximize homology are marked by dashes. Phylogenetically conserved TFBS as well as the CACCC box present only in the mouse PKCδ promoter are indicated (overlined). B, schematic representation of the wild-type or mutated PKCδ promoter reporter constructs containing targeted substitutions in the Sp-binding sites. The potential Sp sites are indicated at the top. The mutated site is marked with ×, and the nonmutated Sp sites are indicated by either circle or square. C, wild-type or mutated reporter constructs as shown in B were individually transfected into NIE115 (black bar) and MN9D (white bar) cells, and luciferase activities were assayed after 24 h. To adjust for transfection efficiency, the plasmid pcDNA3.1-βgal was included in each transfection. The activity of wild-type construct (pGL3−147/+289) was arbitrarily set to 100, and promoter activity of the mutants is expressed as a percentage of the wild-type construct. The results represent the mean ± S.E. of three independent experiments performed in triplicate. The sequences of wild-type and mutated Sp site are shown at the right side of the bar graph. The substituted nucleotides are shown in boldface. D, absence of enhancer elements in the GC-rich sequence (+2/+289) of the mouse PKCδ promoter in NIE115 cells. The PKCδ promoter GC-rich sequence (+2 to +289) was cloned in both orientations into the SalI site of the pGL3−147/+2 reporter constructs as described under “Experimental Procedures.” These constructs were individually transfected into NIE115 cells for 24 h, and luciferase activity was determined. Luciferase activity was normalized with β-galactosidase. The right panel shows the schematic diagram of the constructs. The activity of pGL3−147/+2 was set to 1, and the relative luciferase activity of all other constructs were calculated and expressed as fold of pGL3−147/+2. The results represent the mean ± S.E. of three independent experiments performed in triplicate.
FIGURE 4.
PKCδ promoter activity is stimulated by Sp family members of transcription factors in NIE115 and MN9D cells. A, variable amounts (μg) of pN3-Sp1, pN3-Sp3 FL, pN3-Sp4, or pcDNA-Sp2 expression plasmid or empty vector (pN3 or pcDNA3.1), as indicated, were cotransfected with the PKCδ promoter reporter construct pGL3−147/+289 into NIE115 (black bar) and MN9D (gray bar) cells. Luciferase activity was measured after 24 h of transfection. The plasmid pcDNA3.1-βgal was included in each transfection for data normalization. Values are expressed as fold induction relative to that obtained from cells transfected with 8 μg of empty vector (EV) and represent the mean ± S.E. of three independent experiments performed in triplicate. Variations in the amount of total DNA were compensated with the corresponding empty vector pN3 or pcDNA3.1. B, overexpression of Sp factors in transfected NIE115 (left panel) and MN9D (right panel) cells was determined by immunoblotting analysis. The cells were transfected with Sp expression plasmids in the same manner as A. Whole cell lysates were prepared 24 h after transfection and immunoblotted for Sp1, Sp3, Sp4, or β-actin (loading control). Both short Sp3 (sSp3) and long Sp3 (lSp3) isoforms are shown. C, expression plasmids pN3-Sp1, pN3-Sp3 FL, pN3-Sp4, and empty vector pN3 were cotransfected along with the PKCδ promoter reporter construct pGL3−147/+289 into NIE115 either alone or in the different combinations, as indicated (μg) below the bar graph. Luciferase activity was determined after 24 h of transfection. Data shown represent the mean ± S.E. of three independent experiments performed in triplicate.
FIGURE 5.
Effects of site-directed mutagenesis of Sp-binding sites on PKCδ promoter activity transactivated by overexpression of Sp transcription factors in NIE115 cells. NIE115 cells were cotransfected with the indicated wild-type or mutated PKCδ reporter constructs and 8 μg of pN3-Sp1, pN3-Sp3 FL, pN3-Sp4, pcDNA-Sp2, or empty vector (EV) pN3 or pcDNA3.1. Luciferase activities were assayed after 24 h. The plasmid pcDNA3.1-βgal was included in each transfection to adjust for transfection efficiency. The activity obtained following cotransfection of the wild-type construct (pGL3−147/+209 or pGL3+165/+289) with empty vector (EV) was arbitrarily set to 100, and all other data are expressed as a percentage thereof. The results represent the mean ± S.E. of three independent experiments performed in triplicate. A, schematic representation of the wild-type PKCδ promoter reporter constructs pGL3−147/+209 and pGL3+165/+289. The potential Sp sites are depicted by either a circle or square. B, NIE115 cells were cotransfected with 4 μg of either wild-type (pGL3−147/+209) or mCACCC mutated luciferase reporter constructs along with 8 μg of the expression plasmids pN3-Sp1, pN3-Sp3 FL, pN3-Sp4, pcDNA-Sp2, or empty vector (pN3 or pcDNA3.1). C, wild-type (pGL3+165/+289) or triple mutated luciferase reporter constructs, as indicated, were cotransfected into NIE115 cells along with the expression plasmids for Sp family members of transcription factors. D, wild-type (pGL3+165/+289) or single mutated luciferase reporter constructs, as indicated, were cotransfected into NIE115 cells along with the pcDNA-Sp2 or empty pcDNA3.1 expression vector.
FIGURE 6.
Effects of overexpression of Sp family members of transcription factors on the PKCδ promoter activity in SL2 cells. A, PKCδ promoter reporter construct pGL3−147/+289 (4 μg) was cotransfected with variable amounts (1–4 μg) of Drosophila expression plasmids for Sp1 (pPac-Sp1), the short isoform of Sp3 (pPac-Sp3), the long isoform of Sp3 (pPac-USp3), the full length of Sp3 (pPac-Sp3FL), Sp4 (pN3-Sp4), or Sp2 (pPac-Sp2) in Drosophila SL2 cells. Luciferase activity was measured after 48 h of transfection. The Drosophila β-gal expression plasmid p97b was included in each transfection for data normalization. Values are expressed as fold induction relative to that obtained from cells transfected with 4 μg of empty vector (pPac0) and represent the mean ± S.E. of three independent experiments performed in triplicate. Variations in the amount of total DNA were compensated with the corresponding empty vector pPac0. B, Drosophila expression plasmids pPac-USp3, pPac-Sp1, pPac-Sp4, and pPac-Sp2 were cotransfected along with 4 μg of PKCδ promoter reporter construct pGL3−147/+289 into SL2 cells either alone or in the different combinations, as indicated (μg) below the bar graph. Variations in the amount of total DNA were compensated with the corresponding empty vector pPac0. Luciferase activity was determined after 48 h of transfection. Transfection efficiency was normalized by β-galactosidase activity. Values are expressed as fold induction relative to that obtained from cells transfected with pPac0 alone and represent the mean ± S.E. of three independent experiments performed in triplicate.
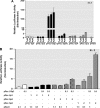
FIGURE 7.
Mithramycin A (MA) inhibits expression of the mouse Prkcd gene. A and B, PKCδ promoter activity is attenuated in NIE115 cells after treatment with mithramycin A. The PKCδ promoter reporter construct pGL3−1694/+289 (A) or pGL3−147/+289 (B) was transfected into NIE115 cells. After 4 h of transfection, the cells were incubated with or without Sp factor inhibitor mithramycin A at concentrations ranging from 0.05 to 5 μ
m
for 24 h. Cells were then harvested, and luciferase activities were determined. The plasmid pcDNA3.1-βgal was included in each transfection to correct the differences in transfection efficiencies. Values are expressed as a percentage of the activity of control and represent the mean ± S.E. of three independent experiments performed in triplicate. (**, p < 0.01; ***, p < 0.001; between the control and mithramycin A-treated samples.) C and D, endogenous PKCδ mRNA levels are reduced by mithramycin A. NIE115 cells (C) or primary striatal neurons (D) were treated with different concentrations of mithramycin A for 24 h. Real time RT PCR analysis of PKCδ mRNA level was performed. β-Actin mRNA level was served as internal control. Values are expressed as a percentage of the activity of control and represent the mean ± S.E. of three independent experiments performed in triplicate. (*, p < 0.05; **, p < 0.01 compared with the control and mithramycin A-treated samples.) E, left panel, exposure of primary striatal neurons to 5 μ
m
mithramycin A reduced PKCδ immunoreactivity. Primary striatal cultures were treated with or without 5 μ
m
MA for 24 h. Cultures were immunostained for PKCδ (red), and the nuclei were counterstained by Hoechst 33342 (blue). Images were obtained using a Nikon TE2000 fluorescence microscope (magnification ×60). Scale bar, 10 μm. Representative immunofluorescence images are shown. The inset shows a higher magnification of the cell body area. Right panel, immunofluorescence quantification of PKCδ fluorescence intensity. Fluorescence immunoreactivity of PKCδ was measured in each group using Metamorph software. Values expressed as percent of control group are mean ± S.E. and representative for results obtained from three separate experiments in triplicate (**, p < 0.01).
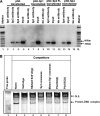
FIGURE 8.
Binding of Sp family of transcription factors to the PKCδ promoter in NIE115 cells. A, ChIP assays in NIE115 cells indicate a physical association of Sp1, Sp3, and Sp4 with the PKCδ promoter region. Cross-linked chromatin was isolated from NIE115 cells transfected with the expression plasmids for Sp1 (pN3-Sp1), Sp3 (pN3-Sp3 FL), Sp4 (pN3-Sp4), or the empty vector pN3, as indicated. Isolated chromatin was enzymatically digested and immunoprecipitated with anti-Sp1 (lanes 2 and 7), anti-Sp3 (lanes 3 and 10), anti-Sp4 (lanes 4 and 13), or antibody-free control (lanes 6, 9, 12, and 15). The subsequently purified DNA from immunoprecipitated samples and unimmunoprecipitated samples (labeled as Input, lanes 5, 8, 11, and 14) was subjected to PCR amplification with primers specific for PKCδ promoter region that generates a 312-bp fragment. B, EMSA to test binding of nuclear proteins from NIE115 cells with the Sp site of the PKCδ promoter. EMSA was performed with an IRye700-labeled probe corresponding to the PKCδ promoter GC(1) and GC(2) motifs and 10 μg of nuclear extract from NIE115 cells. As indicated, various competitors (100-fold excess of unlabeled oligonucleotides, lanes 3–8) were added to the mixture before adding probe. The sequences of the competitors are shown in
supplemental Table S2
. The specific and nonspecific (labeled as N.S.) complexes are indicated by arrows.
FIGURE 9.
PKCδ promoter activity is stimulated by p300/CBP in NIE115 cells, and this effect is independent of p300 HAT activity and requires functional Sp sites. A and B, variable amounts (μg) of expression plasmid for p300 (pCI-p300) and p300 mutant (pCI-p300ΔHAT) (A) or CBP (pcDNA-CBP) (B) were cotransfected with the PKCδ promoter reporter construct pGL3−147/+289 into NIE115 cells. Variations in the amount of total DNA were compensated with the corresponding empty vector (EV) pCIneo or pcDNA3.1. Luciferase activity was measured after 24 h of transfection. The plasmid pcDNA3.1-βgal was included in each transfection for data normalization. Values are expressed as fold induction relative to that obtained from cells transfected with 8 μg of empty vector and represent the mean ± S.E. of three independent experiments performed in triplicate. (**, p < 0.01; ***, p < 0.001; as compared with the EV-transfected samples.) C and D, luciferase reporter constructs Sp1-Luc or mSp1-Luc were cotransfected with variable amounts (μg) of expression plasmid pCI-p300 (C) or pcDNA-CBP (D) were into NIE115 cells. Luciferase activity was measured after 24 h of transfection. Values are expressed as percent of that obtained from cells cotransfected with 8 μg of EV and wild-type Sp1-Luc construct and represent the mean ± S.E. of three independent experiments performed in triplicate.
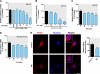
FIGURE 10.
Overexpression of PKCδ sensitizes MN9D dopaminergic cells to oxidative stress-dependent neurodegeneration. MN9D cells were transfected with plasmid expressing PKCδ-GFP or control plasmid EGFP-C1 for 18 h. The cells were then switched to a serum-free medium and exposed to various doses of H2O2, ranging from 0.5 to 2.0 m
m
for 20 h. Cells were collected and assayed for DNA fragmentation (left panel). Data shown represent mean ± S.E. from two independent experiments performed in quadruplicate (*, p < 0.05; **, p < 0.01; ***, p < 0.001; compared with the control and H2O2-treated samples). The overexpression of PKCδ-GFP was confirmed by GFP fluorescence imaging (right panel). Images were obtained using a Nikon TE2000 fluorescence microscope (magnification ×20). Black and white GFP fluorescence images were presented. Scale bar, 100 μm.
FIGURE 11.
Treatment with H2O2 increases Thr505 phosphorylation of PKCδ without affecting PKCδ expression. MN9D cells were incubated with 0.5 m
m
H2O2 for various time spans (3–24 h) and then were harvested for measurement of native PKCδ (A) or PKCδ Thr505 phosphorylation (B) levels by Western blot analysis. β-Actin was used as a loading control. Representative immunoblots are shown. con, control.
FIGURE 12.
Inhibition of PKCδ transcription ameliorates oxidative stress-induced cell death in a dopaminergic neuronal model. A–C, pretreatment with MA inhibited the oxidative stress response in MN9D dopaminergic cells. The cells were preincubated with varying doses of MA for 12 h before treatment with H2O2 (1 m
m
) and then assayed for cell death (A and B) and caspase-3 activity (C). Cell death was measured using the SYTOX® Green cytotoxicity assay as described under “Experimental Procedures.” Representative phase contrast (top) and SYTOX® Green staining (bottom) images are shown. Images were obtained using a Nikon TE2000 fluorescence microscope (magnification ×20). The caspase-3 activity or cytotoxicity was determined and expressed as a percentage of induction relative to unstimulated controls (Con). The results represent the mean ± S.E. of two independent experiments performed in pentaplicate (*, p < 0.05; ***, p < 0.001; as compared with the samples treated with H2O2 alone). D, overexpression of dominant-negative mutant Sp3 protein (pN3-DN-Sp3) lacking the transactivation domains did not enhance the H2O2-induced cell death in MN9D cells. The cells were transfected with indicated amounts of constructs encoding full-length Sp3 (pN3-Sp3 FL), dominant-negative Sp3 (pN3-DN-Sp3), or vehicle vector (pN3) for 16 h. Cells were then exposed to 0.5 m
m
H2O2 for 20 h, and H2O2-induced cell death was determined using the SYTOX® Green cytotoxicity assay. Variations in the amount of total DNA were compensated with the empty vector pN3. The results represent the mean ± S.E. of two independent experiments performed in pentaplicate (**, p < 0.01; between the H2O2-treated samples that were transfected with pN3 or pN3-Sp3 FL vectors).
Similar articles
- Histone hyperacetylation up-regulates protein kinase Cδ in dopaminergic neurons to induce cell death: relevance to epigenetic mechanisms of neurodegeneration in Parkinson disease.
Jin H, Kanthasamy A, Harischandra DS, Kondru N, Ghosh A, Panicker N, Anantharam V, Rana A, Kanthasamy AG. Jin H, et al. J Biol Chem. 2014 Dec 12;289(50):34743-67. doi: 10.1074/jbc.M114.576702. Epub 2014 Oct 23. J Biol Chem. 2014. PMID: 25342743 Free PMC article. - α-Synuclein negatively regulates protein kinase Cδ expression to suppress apoptosis in dopaminergic neurons by reducing p300 histone acetyltransferase activity.
Jin H, Kanthasamy A, Ghosh A, Yang Y, Anantharam V, Kanthasamy AG. Jin H, et al. J Neurosci. 2011 Feb 9;31(6):2035-51. doi: 10.1523/JNEUROSCI.5634-10.2011. J Neurosci. 2011. PMID: 21307242 Free PMC article. - Androgens regulate protein kinase Cdelta transcription and modulate its apoptotic function in prostate cancer cells.
Gavrielides MV, Gonzalez-Guerrico AM, Riobo NA, Kazanietz MG. Gavrielides MV, et al. Cancer Res. 2006 Dec 15;66(24):11792-801. doi: 10.1158/0008-5472.CAN-06-1139. Cancer Res. 2006. PMID: 17178875 - Role of protein kinase Cδ in dopaminergic neurotoxic events.
Shin EJ, Hwang YG, Sharma N, Tran HQ, Dang DK, Jang CG, Jeong JH, Nah SY, Nabeshima T, Kim HC. Shin EJ, et al. Food Chem Toxicol. 2018 Nov;121:254-261. doi: 10.1016/j.fct.2018.09.005. Epub 2018 Sep 6. Food Chem Toxicol. 2018. PMID: 30195712 Review. - Role of proteolytic activation of protein kinase Cdelta in oxidative stress-induced apoptosis.
Kanthasamy AG, Kitazawa M, Kanthasamy A, Anantharam V. Kanthasamy AG, et al. Antioxid Redox Signal. 2003 Oct;5(5):609-20. doi: 10.1089/152308603770310275. Antioxid Redox Signal. 2003. PMID: 14580317 Review.
Cited by
- Striatal molecular signature of subchronic subthalamic nucleus high frequency stimulation in parkinsonian rat.
Lortet S, Lacombe E, Boulanger N, Rihet P, Nguyen C, Kerkerian-Le Goff L, Salin P. Lortet S, et al. PLoS One. 2013 Apr 4;8(4):e60447. doi: 10.1371/journal.pone.0060447. Print 2013. PLoS One. 2013. PMID: 23593219 Free PMC article. - New considerations on hormetic response against oxidative stress.
Luna-López A, González-Puertos VY, López-Diazguerrero NE, Königsberg M. Luna-López A, et al. J Cell Commun Signal. 2014 Dec;8(4):323-31. doi: 10.1007/s12079-014-0248-4. Epub 2014 Oct 5. J Cell Commun Signal. 2014. PMID: 25284448 Free PMC article. - Role of proteolytic activation of protein kinase Cδ in the pathogenesis of prion disease.
Harischandra DS, Kondru N, Martin DP, Kanthasamy A, Jin H, Anantharam V, Kanthasamy AG. Harischandra DS, et al. Prion. 2014 Jan-Feb;8(1):143-53. doi: 10.4161/pri.28369. Prion. 2014. PMID: 24576946 Free PMC article. - Proteolytic activation of proapoptotic kinase protein kinase Cδ by tumor necrosis factor α death receptor signaling in dopaminergic neurons during neuroinflammation.
Gordon R, Anantharam V, Kanthasamy AG, Kanthasamy A. Gordon R, et al. J Neuroinflammation. 2012 Apr 27;9:82. doi: 10.1186/1742-2094-9-82. J Neuroinflammation. 2012. PMID: 22540228 Free PMC article.
References
- Dempsey E. C., Newton A. C., Mochly-Rosen D., Fields A. P., Reyland M. E., Insel P. A., Messing R. O. (2000) Am. J. Physiol. Lung Cell. Mol. Physiol. 279, L429–L438 - PubMed
- Brodie C., Blumberg P. M. (2003) Apoptosis 8, 19–27 - PubMed
- Saito N. (1995) Nippon Yakurigaku Zasshi 105, 127–136 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 NS065167/NS/NINDS NIH HHS/United States
- GM055835/GM/NIGMS NIH HHS/United States
- R01 GM055835/GM/NIGMS NIH HHS/United States
- R01 ES010586/ES/NIEHS NIH HHS/United States
- NS065167/NS/NINDS NIH HHS/United States
- ES10586/ES/NIEHS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Miscellaneous