A diverse range of gene products are effectors of the type I interferon antiviral response - PubMed (original) (raw)
. 2011 Apr 28;472(7344):481-5.
doi: 10.1038/nature09907. Epub 2011 Apr 10.
Affiliations
- PMID: 21478870
- PMCID: PMC3409588
- DOI: 10.1038/nature09907
A diverse range of gene products are effectors of the type I interferon antiviral response
John W Schoggins et al. Nature. 2011.
Erratum in
- Corrigendum: A diverse range of gene products are effectors of the type I interferon antiviral response.
Schoggins JW, Wilson SJ, Panis M, Murphy MY, Jones CT, Bieniasz P, Rice CM. Schoggins JW, et al. Nature. 2015 Sep 3;525(7567):144. doi: 10.1038/nature14554. Epub 2015 Jul 8. Nature. 2015. PMID: 26153858 No abstract available.
Abstract
The type I interferon response protects cells against invading viral pathogens. The cellular factors that mediate this defence are the products of interferon-stimulated genes (ISGs). Although hundreds of ISGs have been identified since their discovery more than 25 years ago, only a few have been characterized with respect to antiviral activity. For most ISG products, little is known about their antiviral potential, their target specificity and their mechanisms of action. Using an overexpression screening approach, here we show that different viruses are targeted by unique sets of ISGs. We find that each viral species is susceptible to multiple antiviral genes, which together encompass a range of inhibitory activities. To conduct the screen, more than 380 human ISGs were tested for their ability to inhibit the replication of several important human and animal viruses, including hepatitis C virus, yellow fever virus, West Nile virus, chikungunya virus, Venezuelan equine encephalitis virus and human immunodeficiency virus type-1. Broadly acting effectors included IRF1, C6orf150 (also known as MB21D1), HPSE, RIG-I (also known as DDX58), MDA5 (also known as IFIH1) and IFITM3, whereas more targeted antiviral specificity was observed with DDX60, IFI44L, IFI6, IFITM2, MAP3K14, MOV10, NAMPT (also known as PBEF1), OASL, RTP4, TREX1 and UNC84B (also known as SUN2). Combined expression of pairs of ISGs showed additive antiviral effects similar to those of moderate type I interferon doses. Mechanistic studies uncovered a common theme of translational inhibition for numerous effectors. Several ISGs, including ADAR, FAM46C, LY6E and MCOLN2, enhanced the replication of certain viruses, highlighting another layer of complexity in the highly pleiotropic type I interferon system.
Figures
Figure 1. FACS-based screen for identifying antiviral ISGs
a. Gateway-compatible bicistronic lentiviral vector. b. Schematic of overexpression screen showing hypothetical FACS plots of inhibitory versus non-inhibitory genes. Overlap of RFP (magenta) and GFP (green) is depicted as white. c. FACS plots showing _IFI6_-mediated inhibition of YFV in _STAT1_−/−Fib. d. Distribution of HCV replication levels at 48 h normalized to Fluc control. The number of ISGs within standard deviation (S.D. or Z-score) ranges are shown in boxes. e. Dot plots of HCV replication levels at 48 h and 72 h normalized to Fluc control. Select ISGs are denoted in blue. Black line indicates population mean.
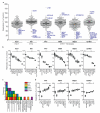
Figure 2. Identification of ISGs that inhibit or enhance virus replication
a. Dot plots of large-scale ISG screens against six viruses. Replication levels were normalized to Fluc control. Select ISGs are denoted in blue. Black line indicates population mean. b, d. Confirmation assays of selected ISGs. For HIV, Macaca mulatta (M.m.) TRIM5 was included as a control. Replication levels were normalized to Fluc control. Data are represented by box and whisker plots. n=8 for HCV, n=9 for other viruses. Statistical significance was determined by one-way ANOVA. (***,P<0.001, **,P<0.01, *,P<0.05, ns-not significant). c. Frequency of validated antiviral ISG prevalence across six screens.
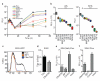
Figure 3. Combinatorial action of inhibitory and enhancing ISGs
a. Anti-HCV ISGs were tested in 2-gene combinations (inset). Dashed red line denotes the strongest single-gene inhibitor, IRF1. b, d, e. Confirmation assays from two-gene screens for HCV (b), HIV(d), YFV(e). Data were normalized to Fluc control. n=8 for HCV, YFV, n=9 for HIV. Statistical significance was determined by one-way ANOVA. (***,P<0.001, **,P<0.01, *,P<0.05, ns-not significant). Inhibition of HIV IFNα is plotted for comparison. ISGs are color-coded: control (red), inhibitory (black), enhancing (green) c. Inhibition of HCV by IFNβ. Results are mean ± s.d., n=3.
Figure 4. Translational inhibition as a common mechanism of ISG-mediated antiviral action
a. ISG-expressing Huh-7 cells were transfected with HCV subgenomic replicon RNA and Gluc levels in cell supernatants were measured b. Inhibition of primary translation (left panel) or replication (right panel) was inferred from 4 h and 72 h data, respectively. Results are mean ± s.d., n=4. Statistical significance was determined by one-way ANOVA. (***,P<0.001, **,P<0.01, *,P<0.05, ns-not significant) d-g. C6orf150-mediated inhibition of SINV in _STAT1_−/−Fib. C6orf150 inhibits SINV-GFP replication (d) and non-GFP SINV production(e). C6orf150 and IRF1 inhibit SINV(ts6) primary translation (f) and SINV-Fluc replication (g). Results are mean ± s.d., n=3.
Comment in
- A dive into the complexity of type I interferon antiviral functions.
Touzot M, Soumelis V, Asselah T. Touzot M, et al. J Hepatol. 2012 Mar;56(3):726-8. doi: 10.1016/j.jhep.2011.07.009. Epub 2011 Jul 27. J Hepatol. 2012. PMID: 21801700 No abstract available. - Large-scale identification of effector genes that mediate the type I interferon antiviral response.
Takahashi K, Marusawa H, Chiba T. Takahashi K, et al. Gastroenterology. 2012 Jan;142(1):178-80. doi: 10.1053/j.gastro.2011.11.008. Epub 2011 Nov 19. Gastroenterology. 2012. PMID: 22107719 No abstract available.
Similar articles
- Interferon-stimulated genes and their antiviral effector functions.
Schoggins JW, Rice CM. Schoggins JW, et al. Curr Opin Virol. 2011 Dec;1(6):519-25. doi: 10.1016/j.coviro.2011.10.008. Curr Opin Virol. 2011. PMID: 22328912 Free PMC article. Review. - The Interferon-Induced Exonuclease ISG20 Exerts Antiviral Activity through Upregulation of Type I Interferon Response Proteins.
Weiss CM, Trobaugh DW, Sun C, Lucas TM, Diamond MS, Ryman KD, Klimstra WB. Weiss CM, et al. mSphere. 2018 Sep 19;3(5):e00209-18. doi: 10.1128/mSphere.00209-18. mSphere. 2018. PMID: 30232164 Free PMC article. - IRF-1, RIG-I and MDA5 display potent antiviral activities against norovirus coordinately induced by different types of interferons.
Dang W, Xu L, Yin Y, Chen S, Wang W, Hakim MS, Chang KO, Peppelenbosch MP, Pan Q. Dang W, et al. Antiviral Res. 2018 Jul;155:48-59. doi: 10.1016/j.antiviral.2018.05.004. Epub 2018 May 10. Antiviral Res. 2018. PMID: 29753657 - Two Interferon-Stimulated Response Elements Cooperatively Regulate Interferon-Stimulated Gene Expression in West Nile Virus-Infected IFNAR-/- Mouse Embryo Fibroblasts.
Cui D, Espínola EE, Arora K, Brinton MA. Cui D, et al. J Virol. 2021 Oct 27;95(22):e0104021. doi: 10.1128/JVI.01040-21. Epub 2021 Sep 8. J Virol. 2021. PMID: 34495694 Free PMC article. - Fish interferon-stimulated genes: The antiviral effectors.
Poynter SJ, DeWitte-Orr SJ. Poynter SJ, et al. Dev Comp Immunol. 2016 Dec;65:218-225. doi: 10.1016/j.dci.2016.07.011. Epub 2016 Jul 19. Dev Comp Immunol. 2016. PMID: 27451256 Review.
Cited by
- Crosstalk Between SUMO and Ubiquitin-Like Proteins: Implication for Antiviral Defense.
Chelbi-Alix MK, Thibault P. Chelbi-Alix MK, et al. Front Cell Dev Biol. 2021 Apr 21;9:671067. doi: 10.3389/fcell.2021.671067. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 33968942 Free PMC article. Review. - Sequence-Specific Modifications Enhance the Broad-Spectrum Antiviral Response Activated by RIG-I Agonists.
Chiang C, Beljanski V, Yin K, Olagnier D, Ben Yebdri F, Steel C, Goulet ML, DeFilippis VR, Streblow DN, Haddad EK, Trautmann L, Ross T, Lin R, Hiscott J. Chiang C, et al. J Virol. 2015 Aug;89(15):8011-25. doi: 10.1128/JVI.00845-15. Epub 2015 May 27. J Virol. 2015. PMID: 26018150 Free PMC article. - Systems analysis of a RIG-I agonist inducing broad spectrum inhibition of virus infectivity.
Goulet ML, Olagnier D, Xu Z, Paz S, Belgnaoui SM, Lafferty EI, Janelle V, Arguello M, Paquet M, Ghneim K, Richards S, Smith A, Wilkinson P, Cameron M, Kalinke U, Qureshi S, Lamarre A, Haddad EK, Sekaly RP, Peri S, Balachandran S, Lin R, Hiscott J. Goulet ML, et al. PLoS Pathog. 2013;9(4):e1003298. doi: 10.1371/journal.ppat.1003298. Epub 2013 Apr 25. PLoS Pathog. 2013. PMID: 23633948 Free PMC article. - Pre-infection antiviral innate immunity contributes to sex differences in SARS-CoV-2 infection.
Sauerwald N, Zhang Z, Ramos I, Nair VD, Soares-Schanoski A, Ge Y, Mao W, Alshammary H, Gonzalez-Reiche AS, van de Guchte A, Goforth CW, Lizewski RA, Lizewski SE, Amper MAS, Vasoya M, Seenarine N, Guevara K, Marjanovic N, Miller CM, Nudelman G, Schilling MA, Sealfon RSG, Termini MS, Vangeti S, Weir DL, Zaslavsky E, Chikina M, Wu YN, Van Bakel H, Letizia AG, Sealfon SC, Troyanskaya OG. Sauerwald N, et al. Cell Syst. 2022 Nov 16;13(11):924-931.e4. doi: 10.1016/j.cels.2022.10.005. Epub 2022 Nov 1. Cell Syst. 2022. PMID: 36323307 Free PMC article. - Differential expression of HIV-1 interfering factors in monocyte-derived macrophages stimulated with polarizing cytokines or interferons.
Cobos Jiménez V, Booiman T, de Taeye SW, van Dort KA, Rits MA, Hamann J, Kootstra NA. Cobos Jiménez V, et al. Sci Rep. 2012;2:763. doi: 10.1038/srep00763. Epub 2012 Oct 23. Sci Rep. 2012. PMID: 23094138 Free PMC article.
References
References for Print Article
- de Veer MJ, et al. Functional classification of interferon-stimulated genes identified using microarrays. J Leukoc Biol. 2001;69:912–920. - PubMed
- Dupuis S, et al. Impaired response to interferon-alpha/beta and lethal viral disease in human STAT1 deficiency. Nat Genet. 2003;33:388–391. - PubMed
- Keskinen P, et al. Impaired antiviral response in human hepatoma cells. Virology. 1999;263:364–375. - PubMed
References for Supplemental Information
- Tsetsarkin K, et al. Infectious clones of Chikungunya virus (La Reunion isolate) for vector competence studies. Vector Borne Zoonotic Dis. 2006;6:325–337. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- AI064003/AI/NIAID NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- F32 DK081193/DK/NIDDK NIH HHS/United States
- AI057158/AI/NIAID NIH HHS/United States
- R37 AI064003/AI/NIAID NIH HHS/United States
- R01 AI064003/AI/NIAID NIH HHS/United States
- F32 DK082155/DK/NIDDK NIH HHS/United States
- DK081193/DK/NIDDK NIH HHS/United States
- F32 DK081193-01A1/DK/NIDDK NIH HHS/United States
- U54 AI057158/AI/NIAID NIH HHS/United States
- F32 DK082155-01/DK/NIDDK NIH HHS/United States
- R01 AI064003-01/AI/NIAID NIH HHS/United States
- DK082155/DK/NIDDK NIH HHS/United States
- U54 AI057158-01/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous