A framework for variation discovery and genotyping using next-generation DNA sequencing data - PubMed (original) (raw)
Comparative Study
doi: 10.1038/ng.806. Epub 2011 Apr 10.
Eric Banks, Ryan Poplin, Kiran V Garimella, Jared R Maguire, Christopher Hartl, Anthony A Philippakis, Guillermo del Angel, Manuel A Rivas, Matt Hanna, Aaron McKenna, Tim J Fennell, Andrew M Kernytsky, Andrey Y Sivachenko, Kristian Cibulskis, Stacey B Gabriel, David Altshuler, Mark J Daly
Affiliations
- PMID: 21478889
- PMCID: PMC3083463
- DOI: 10.1038/ng.806
Comparative Study
A framework for variation discovery and genotyping using next-generation DNA sequencing data
Mark A DePristo et al. Nat Genet. 2011 May.
Abstract
Recent advances in sequencing technology make it possible to comprehensively catalog genetic variation in population samples, creating a foundation for understanding human disease, ancestry and evolution. The amounts of raw data produced are prodigious, and many computational steps are required to translate this output into high-quality variant calls. We present a unified analytic framework to discover and genotype variation among multiple samples simultaneously that achieves sensitive and specific results across five sequencing technologies and three distinct, canonical experimental designs. Our process includes (i) initial read mapping; (ii) local realignment around indels; (iii) base quality score recalibration; (iv) SNP discovery and genotyping to find all potential variants; and (v) machine learning to separate true segregating variation from machine artifacts common to next-generation sequencing technologies. We here discuss the application of these tools, instantiated in the Genome Analysis Toolkit, to deep whole-genome, whole-exome capture and multi-sample low-pass (∼4×) 1000 Genomes Project datasets.
Figures
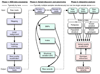
Figure 1
Framework for variation discovery and genotyping from next-generation DNA sequencing. See text for a detailed description.
Figure 2
IGV visualization of alignments in region chr1:1,510,446–1,510,622 from the (a) Trio NA12878 Illumina reads from 1000 Genomes and (b) NA12878 HiSeq reads before (left) and after (right) multiple sequence realignment. Reads are depicted as arrows oriented by increasing machine cycle; highlighted bases indicate mismatches to the reference: A is green, G is orange, T is red, and deleted bases are dashes; a coverage histogram per base is shown above the reads. Both the 4bp indel (rs34877486) and the C/T polymorphism (rs28788874) are present in dbSNP, as are the artifactual A/G polymorphisms (rs28782535 and rs28783181) resulting from the mis-modeled indel, indicating that these sites are common misalignment errors.
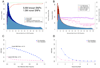
Figure 3
Raw (violet) and recalibrated (blue) base quality scores for NGS paired end read sets of NA12878 of (a) Illumina/GA (b) Life/SOLiD and (c) Roche/454 lanes from 1000 Genomes, and (d) Illumina/HiSeq. For each technology: top panel: shows reported base quality scores compared to the empirical estimates (Methods); middle panel: the difference between the average reported and empirical quality score for each machine cycle, with positive and negative cycle values given for the first and second read in the pair, respectively; bottom panel: the difference between reported and empirical quality scores for each of the 16 genomic dinucleotide contexts. For example, the AG context occurs at all sites in a read where G is the current nucleotide and A is the preceding one in the read. Root-mean-square errors (RMSE) are given for the pre- and post-recalibration curves.
Figure 4
(a) Relationship in the HiSeq call set between strand bias and quality by depth, for genomic locations in HapMap3 (red) and dbSNP (yellow) used for training the variant quality score recalibrator (left) and the same annotations applied to differentiate likely true positive (green) from false positive (purple) novel SNPs. (b,c,d) Quality tranches in the recalibrated HiSeq (b), exome (c), and low-pass CEU (d) calls beginning with (top) the highest-quality but smallest call set with an estimated false positive rate among novel SNP calls of <1/1000 to a more comprehensive call set (bottom) that includes effectively all true positives in the raw call set along with more false positive calls for a cumulative false positive rate of 10%. Each successive call set contains within it the previous tranche’s true and false positive calls (shaded bars) as well as tranche-specific calls of both classes (solid bars). The tranche selected for further analyses here is indicated.
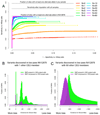
Figure 5
Variation discovered among 60 individuals from the CEPH population from 1000 Genomes pilot phase plus low-pass NA12878. (a) Discovered SNPs by non-reference allele count in the 61 CEPH cohort, colored by known (light blue, striped) and novel (dark blue, filled) variation, along with non-reference sensitivity to CEU HapMap3 and 1000 Genomes low-pass variants. (b) Quality and certainty of discovered SNPs by non-reference allele count. The histogram depicts the certainty of called variation broken out into 0.1, 1, and 10% novel FDR tranches. The Ti/Tv ratio is shown for known and novel variation for each allele count, aggregating the novel calls with allele count > 74 due to their limited numbers. (c,d) Genotyping accuracy for NA12878 from reads alone (blue circles) and following genotype-likelihood based imputation (pink squares) called in the 61 sample call set as assessed by the NRD rate to HiSeq genotypes, as a function of allele count (c) and sequencing depth (d).
Figure 6
Sensitivity and specificity of multi-sample discovery of variation in NA12878 with increasing cohort size for low-pass NA12878 read sets processed with N additional CEPH samples. (a) Receiver operating characteristic (ROC) curves for SNP calls relating specificity and sensitivity to discover non-reference sites from the NA12878 HiSeq call set. The maximum callable sensitivity, 66%, is the percent of sites from the HiSeq call set where at least one read carries the alternate allele in the low-pass data for NA12878; it reflects both differences in the sequencing technologies (36–76bp GAII for the low-pass NA12878 sample vs. 101bp HiSeq) as well as the vagaries of sampling at 4× coverage. Because most of these missed sites are common and are consequently called in the other samples, imputation recovers ~50% of these sites. (b,c) Increasing power to identify strand-biased, likely false positive SNP calls with additional samples. Histograms of the Strand Bias annotation at raw variant calls discovered in the low-pass CEU data using NA12878 at 4× combined with one other CEU individual (b) and with 60 other individuals (c) stratified into sites present (green) and not (purple) in the 1000 Genomes CEU trio.
Similar articles
- An efficient and scalable analysis framework for variant extraction and refinement from population-scale DNA sequence data.
Jun G, Wing MK, Abecasis GR, Kang HM. Jun G, et al. Genome Res. 2015 Jun;25(6):918-25. doi: 10.1101/gr.176552.114. Epub 2015 Apr 16. Genome Res. 2015. PMID: 25883319 Free PMC article. - A probabilistic method for the detection and genotyping of small indels from population-scale sequence data.
Bansal V, Libiger O. Bansal V, et al. Bioinformatics. 2011 Aug 1;27(15):2047-53. doi: 10.1093/bioinformatics/btr344. Epub 2011 Jun 7. Bioinformatics. 2011. PMID: 21653520 Free PMC article. - A survey of tools for variant analysis of next-generation genome sequencing data.
Pabinger S, Dander A, Fischer M, Snajder R, Sperk M, Efremova M, Krabichler B, Speicher MR, Zschocke J, Trajanoski Z. Pabinger S, et al. Brief Bioinform. 2014 Mar;15(2):256-78. doi: 10.1093/bib/bbs086. Epub 2013 Jan 21. Brief Bioinform. 2014. PMID: 23341494 Free PMC article. - Model-based quality assessment and base-calling for second-generation sequencing data.
Bravo HC, Irizarry RA. Bravo HC, et al. Biometrics. 2010 Sep;66(3):665-74. doi: 10.1111/j.1541-0420.2009.01353.x. Biometrics. 2010. PMID: 19912177 Free PMC article. Review. - Genome structural variation discovery and genotyping.
Alkan C, Coe BP, Eichler EE. Alkan C, et al. Nat Rev Genet. 2011 May;12(5):363-76. doi: 10.1038/nrg2958. Epub 2011 Mar 1. Nat Rev Genet. 2011. PMID: 21358748 Free PMC article. Review.
Cited by
- Whole-exome profiles of inflammatory breast cancer and pathological response to neoadjuvant chemotherapy.
Bertucci F, Guille A, Lerebours F, Ceccarelli M, Syed N, Adélaïde J, Finetti P, Ueno NT, Van Laere S, Viens P, De Nonneville A, Goncalves A, Birnbaum D, Callens C, Bedognetti D, Mamessier E. Bertucci F, et al. J Transl Med. 2024 Oct 27;22(1):969. doi: 10.1186/s12967-024-05790-8. J Transl Med. 2024. PMID: 39465437 Free PMC article. - A multiplex PCR amplicon sequencing assay to screen genetic hearing loss variants in newborns.
Yang H, Luo H, Zhang G, Zhang J, Peng Z, Xiang J. Yang H, et al. BMC Med Genomics. 2021 Feb 27;14(1):61. doi: 10.1186/s12920-021-00906-1. BMC Med Genomics. 2021. PMID: 33639928 Free PMC article. - Combined immunodeficiency due to MALT1 mutations, treated by hematopoietic cell transplantation.
Punwani D, Wang H, Chan AY, Cowan MJ, Mallott J, Sunderam U, Mollenauer M, Srinivasan R, Brenner SE, Mulder A, Claas FH, Weiss A, Puck JM. Punwani D, et al. J Clin Immunol. 2015 Feb;35(2):135-46. doi: 10.1007/s10875-014-0125-1. Epub 2015 Jan 28. J Clin Immunol. 2015. PMID: 25627829 Free PMC article. - Reduced Representation Libraries from DNA Pools Analysed with Next Generation Semiconductor Based-Sequencing to Identify SNPs in Extreme and Divergent Pigs for Back Fat Thickness.
Bovo S, Bertolini F, Schiavo G, Mazzoni G, Dall'Olio S, Fontanesi L. Bovo S, et al. Int J Genomics. 2015;2015:950737. doi: 10.1155/2015/950737. Epub 2015 Mar 4. Int J Genomics. 2015. PMID: 25821781 Free PMC article. - The use of museum specimens with high-throughput DNA sequencers.
Burrell AS, Disotell TR, Bergey CM. Burrell AS, et al. J Hum Evol. 2015 Feb;79:35-44. doi: 10.1016/j.jhevol.2014.10.015. Epub 2014 Dec 18. J Hum Evol. 2015. PMID: 25532801 Free PMC article.
References
- Lee W, et al. The mutation spectrum revealed by paired genome sequences from a lung cancer patient. Nature. 2010;465:473–477. - PubMed
Publication types
MeSH terms
Grants and funding
- P30 DK043351/DK/NIDDK NIH HHS/United States
- U01 HG005208/HG/NHGRI NIH HHS/United States
- U54 HG003067/HG/NHGRI NIH HHS/United States
- 54 HG003067/HG/NHGRI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases