Nonreceptor tyrosine kinase BMX maintains self-renewal and tumorigenic potential of glioblastoma stem cells by activating STAT3 - PubMed (original) (raw)
. 2011 Apr 12;19(4):498-511.
doi: 10.1016/j.ccr.2011.03.004.
Qiulian Wu, Lin Cheng, Justin D Lathia, Zhi Huang, Jinbo Yang, Jennifer MacSwords, Christine E Eyler, Roger E McLendon, John M Heddleston, Weinian Shou, Dolores Hambardzumyan, Jeongwu Lee, Anita B Hjelmeland, Andrew E Sloan, Markus Bredel, George R Stark, Jeremy N Rich, Shideng Bao
Affiliations
- PMID: 21481791
- PMCID: PMC3076106
- DOI: 10.1016/j.ccr.2011.03.004
Nonreceptor tyrosine kinase BMX maintains self-renewal and tumorigenic potential of glioblastoma stem cells by activating STAT3
Olga A Guryanova et al. Cancer Cell. 2011.
Abstract
Glioblastomas display cellular hierarchies containing tumor-propagating glioblastoma stem cells (GSCs). STAT3 is a critical signaling node in GSC maintenance but molecular mechanisms underlying STAT3 activation in GSCs are poorly defined. Here we demonstrate that the bone marrow X-linked (BMX) nonreceptor tyrosine kinase activates STAT3 signaling to maintain self-renewal and tumorigenic potential of GSCs. BMX is differentially expressed in GSCs relative to nonstem cancer cells and neural progenitors. BMX knockdown potently inhibited STAT3 activation, expression of GSC transcription factors, and growth of GSC-derived intracranial tumors. Constitutively active STAT3 rescued the effects of BMX downregulation, supporting that BMX signals through STAT3 in GSCs. These data demonstrate that BMX represents a GSC therapeutic target and reinforces the importance of STAT3 signaling in stem-like cancer phenotypes.
Copyright © 2011 Elsevier Inc. All rights reserved.
Figures
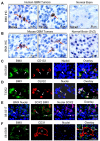
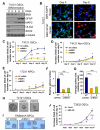
Figure 1. Expression of BMX in a Subpopulation of Cancer Cells Expressing Stem Cell Markers in GBMs
(A) Immunohistochemical (IHC) staining of BMX in human primary GBMs and normal brain tissues. BMX(+) cells (indicated by arrows) are shown in brown. Sections were counterstained with hematoxylin to show nuclei. (B) IHC staining of BMX in GBMs from a genetically engineered mouse model and normal mouse brains including the subventricular zone (SVZ). BMX(+) cells (brown) are indicated by arrows. (C-E) Immunofluorescent (IF) staining of BMX and the GSC markers CD133 (C), OLIG2 (D) and SOX2 (E) on frozen sections of GBMs. BMX was labeled in green, CD133, OLIG2 or SOX2 in red. Nuclei were counterstained with DAPI (blue). Cells with extensive overlap are marked with arrows. (F) IF staining of BMX(+) cells (green) in relation to blood vessels marked by CD31 staining for endothelial cells (red) in GBMs. Nuclei were counterstained with DAPI (blue). Vessel lumen was denoted by “*”. See also Figure S1, and Tables S1 and S2.
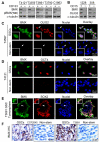
Figure 2. Differential Expression of BMX in GSCs Relative to Non-stem Tumor Cells
(A) Immunoblot (IB) analysis of BMX protein and its activating phosphorylation [pBMX(Y40)] in CD133-enriched GSCs and matched CD133-depleted tumor cells derived from five GBMs. (B) IB analysis of BMX and the GSC marker OLIG2 in CD15-enriched GSCs and CD15-depleted tumor cells from two GBMs. (C-E) IF staining of BMX with several GSC markers including OLIG2 (C), OCT4 (D) and SOX2 (E) in GSCs. BMX was labeled in green, OLIG2, OCT4 or SOX2 in red; and nuclei were counterstained with DAPI (blue). GSCs were cultured as attached monolayers (C and D) or tumorspheres (E). (F) IHC staining of BMX in cultured GSCs and matched non-stem tumor cells (Non-stem) from a primary GBM (CCF2045) and a xenograft (T3691). See also Figure S2.
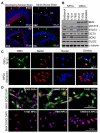
Figure 3. Preferential Expression of BMX in Human GSCs but Not in NPCs and Subventricular Zone (SVZ) of Mouse Brains
(A) IF staining of BMX in SVZs, SEZs (subependimal zones) and DG (dentate gyrus) in developing and adult mouse brains. Mouse brains were stained for BMX (green) and a NPC marker SOX2 (red). Nuclei were counterstained with DAPI (blue). *, ventricles; CC: corpus callosum. (B) IB analyses of BMX, STAT3, pSTAT3(Y705), OCT4, SOX2 and OLIG2 expression in 3 GSC populations and 4 human NPC lines. (C) IF staining of BMX (green) and the NPC marker Nestin (red) in GSCs (T4121) and NPCs (ENStemA). Nuclei were counterstained with DAPI (blue). (D) IF staining of BMX (green) and SOX2 (red) in 3 GSC populations and 3 human NPC lines (bottom panels). Nuclei were counterstained with DAPI (blue). See also Figure S3.
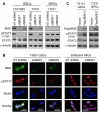
Figure 4. BMX Is Required for STAT3 Activation in GSCs but not in NPCs
(A) IB analysis of activating phosphorylation of STAT3 (Y705) and total STAT3 in GSCs derived from the indicated GBMs and NPCs with BMX knockdown by shBMX1 or shBMX2. NT shRNA: non-targeting control shRNA. (B) IF staining of phosphorylated STAT3 [pSTAT3(Y705)] in GSCs and NPCs with BMX knockdown. BMX was labeled in green, pSTAT3 in red, and nuclei were counterstained with DAPI (blue). (C) IB analysis of STAT3 activating phosphorylation and total STAT3 in GSCs and NPCs with ectopic expression of a constitutively active BMX with a Flag tag (Flag-BMX-C). See also Figure S4.
Figure 5. BMX Declines during GSC Differentiation and BMX Disruption Decreases GSC Proliferation and Tumorsphere Formation
(A) IB analysis of BMX expression during GSC differentiation. Protein levels of BMX, SOX2 and OLIG2 (GSC transcription factors), GFAP (astrocyte marker), and MAP2 (neuronal marker) were examined during GSC differentiation induced by serum (10% FBS) at indicated times. (B) IF staining of BMX (green) and the neuronal marker MAP2 (red) or the astrocyte marker GFAP (red) from day 0 to day 6 during GSC differentiation induced by serum. Nuclei were counterstained with DAPI (blue). (C-E) Effects of BMX knockdown with shBMX1 or shBMX2 on cell proliferation in GSCs (C), matched non-stem tumor cells (D), and human NPCs (17231) (E). Cells (1×104 per well) transduced with shBMX or NT shRNA were plated in quadruplicate wells in stem cell media, and then counted at the indicated times (day 0 to day 12). Data are means ± SD (n=4). *p > 0.05; ***p < 0.001. (F-H) Effects of BMX knockdown by shBMX1 or shBMX2 on GSC tumorsphere formation. Representative images of tumorspheres derived from GSCs expressing NT shRNA, shBMX1 or shBMX2 are shown (H). Quantification shows reduced GSC tumorsphere number (F) and size (G) by shBMXs. Data are means ± SD (n=3). ***p < 0.001. (I) Effect of shBMX1 or shBMX2 on neurosphere formation of NPCs. The representative images of neurospheres derived from NPCs expressing NT shRNA, shBMX1 or shBMX2 are shown. (J) Effects of a dominant-negative BMX (BMX-DN) on cell proliferation of GSCs. Data are means ± SD (n=4). *p < 0.002; ***p < 0.001. See also Figure S5.
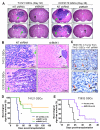
Figure 6. Targeting BMX in GSCs Suppressed Tumor Growth and Increased Survival of Animals Bearing Intracranial GBMs
Effects of targeting BMX on GBM tumor growth and animal survival were examined in the intracranial xenograft model. GSCs from indicated GBMs were transduced with NT shRNA, shBMX1, BMX-DN or vector control through lentiviral infection, and then intracranially transplanted into the brains of immunocompromised mice (5×103 cells per mouse). Mouse brains implanted with GSCs expressing NT shRNA or shBMX were harvested simultaneously to examine the impact of BMX disruption on GBM tumor growth (A-C). In the animal survival experiments (D and E), mice implanted with GSCs expressing NT shRNA, shBMX, BMX-DN or vector control were maintained until the development of neurological signs or for 180 days. (A) Representative images of cross sections (H&E stained) of mouse brains harvested on day 32 (T4121 GSCs) or day 36 (CCF2170 GSCs) after transplantation of GSCs expressing NT shRNA or shBMX. Arrows indicate large tumors in the brains with GSCs expressing NT shRNA, or small tumors in the brains with GSCs expressing shBMX. (B) Histological analysis of brain tumors derived from GSCs expressing NT shRNA or shBMX. Green arrows indicate cancer cells invading into normal tissue in brains implanted with GSCs expressing NT shRNA. A black arrow indicates a small tumor nodule in the brain implanted with GSCs expressing shBMX1. (C) IHC staining of BMX in GBM xenografts derived from GSCs expressing NT shRNA. Arrows indicate the representative BMX(+) cells (brown). Sections were counterstained with hematoxylin. (D) Kaplan-Meier survival curves of mice implanted with GSCs expressing shBMX1, shBMX2 or NT shRNA. *p < 0.001. (E) Kaplan-Meier survival curves of mice implanted with GSCs transfected with a dominant-negative BMX (BMX-DN) or vector control. **p < 0.001. See also Figure S6.
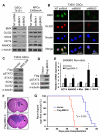
Figure 7. BMX Regulates Expression of Stem Cell Transcription Factors in GSCs but not in NPCs
(A) IB analysis of stem cell transcription factors (SCTFs) in GSCs and NPCs with BMX knockdown by shBMX1 or shBMX2. Cell lysates from the indicated GSCs and NPCs expressing shBMX1, shBMX2 or NT shRNA were analyzed for protein levels of BMX, OLIG2, OCT4, SOX2, and NANOG. (B) IF staining of OLIG2 (a GSC transcription factor) in GSCs expressing NT shRNA, shBMX1 or shBMX2. BMX was labeled in green, OLIG2 in red. Nuclei were counterstained with DAPI (blue). (C) IB analysis of STAT3 activating phosphorylation (pSTAT3-Y705) and expression of GSC transcription factors SOX2 and OLIG2 in GSCs expressing a dominant-negative BMX (BMX-DN). (D) IB analysis of STAT3 activation (pSTAT3-Y705) by expression of a constitutively active BMX (Flag-BMX-C) in D456MG non-stem tumor cells. (E) RT-PCR analysis of mRNA expression of SCTFs including BMI-1, NANOG, OCT4 and KLF4 in D456MG non-stem tumor cells expressing the active Flag-BMX-C. Data are means ± SD (n=3). *p < 0.002. (F) Effect of the active Flag-BMX-C on tumor growth of GBM xenografts derived from D456MG non-stem tumor cells. D456 non-stem tumor cells were transduced with Flag-BMX-C or vector control, and then transplanted into the brains of immuno-deficient mice via intracranial injection (2×106 cells per mouse). The brains were harvested simultaneously on day 42 after cell transplantation. The representative images of brain cross sections (H&E stained) are shown. Tumors inside brains are indicated by arrows. (G) Kaplan-Meier survival curves of mice implanted with D456MG non-stem tumor cells transfected with the active Flag-BMX-C or control vector. D456 non-stem tumor cells transduced with Flag-BMX-C or control vector were intracranially transplanted into brains of immuno-deficient mice (2×106 cells per mouse). The mice were maintained until the development of neurological signs or for 180 days. *p = 0.002 by log-rank survival analysis. See also Figure S7.
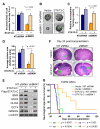
Figure 8. Ectopic Expression of a Constitutively Active STAT3 in GSCs Rescued the Phenotypes Caused by BMX Downregulation
(A) Effects of a constitutively active STAT3 (STAT3-C-Flag) on cell proliferation of GSCs expressing NT shRNA or shBMX. GSCs (CCF2045) were transduced with STAT3-C-Flag or vector control and then targeted with shBMX1 or NT shRNA. The relative increase (folds) of cell number from day 0 to day 4 among the four groups (STAT3-C/Vector and shBMX1/NTshRNA) was compared. Data are means ± SD (n=5). *p < 0.001. (B-D) Effects of STAT3-C-Flag on tumorsphere formation of GSCs expressing NT shRNA or shBMX. Representative images of GSC tumorspheres derived from GSCs transduced with STAT3-C-Flag or vector and targeted with shBMX1 or NT shRNA are shown (B). Quantification shows the effects of STAT3-C-Flag expression on tumorsphere number (C) and size (D) of GSCs expressing NT shRNA or shBMX. Data are means ± SD (n=4). *p<0.001. (E) IB analysis of GSC transcription factors (SOX2, OLIG2, OCT4 and NANOG) in CCF2045 GSCs transduced with STAT3-C-Flag or control vector in combination with shBMX or NT shRNA. (F) Effects of STAT3-C expression on tumor growth of GSCs expressing shBMX or NT shRNA. GSCs (T3359) were transduced with STAT3-C-Flag or vector control, and then infected with shBMX1 or NT shRNA lentiviruses. 48 hours after infection, cells were transplanted into the brains of immunocompromised mice. Representative images of cross sections (H&E stained) of mouse brains from the indicated experimental groups harvested 32 days post-transplantation are shown. Arrows indicate tumors in mouse brains. (G) Kaplan-Meier survival curves of mice implanted with GSCs expressing STAT3-C or vector in combination with shBMX or NT shRNA. Four groups of mice implanted with the GSCs were maintained until the development of neurological signs or for 180 days. The statistical analyses (p value) between each group are shown under the figure.
Similar articles
- Tetraspanin CD9 stabilizes gp130 by preventing its ubiquitin-dependent lysosomal degradation to promote STAT3 activation in glioma stem cells.
Shi Y, Zhou W, Cheng L, Chen C, Huang Z, Fang X, Wu Q, He Z, Xu S, Lathia JD, Ping Y, Rich JN, Bian XW, Bao S. Shi Y, et al. Cell Death Differ. 2017 Jan;24(1):167-180. doi: 10.1038/cdd.2016.110. Epub 2016 Oct 14. Cell Death Differ. 2017. PMID: 27740621 Free PMC article. - Ibrutinib inactivates BMX-STAT3 in glioma stem cells to impair malignant growth and radioresistance.
Shi Y, Guryanova OA, Zhou W, Liu C, Huang Z, Fang X, Wang X, Chen C, Wu Q, He Z, Wang W, Zhang W, Jiang T, Liu Q, Chen Y, Wang W, Wu J, Kim L, Gimple RC, Feng H, Kung HF, Yu JS, Rich JN, Ping YF, Bian XW, Bao S. Shi Y, et al. Sci Transl Med. 2018 May 30;10(443):eaah6816. doi: 10.1126/scitranslmed.aah6816. Sci Transl Med. 2018. PMID: 29848664 Free PMC article. - TRIM8 regulates stemness in glioblastoma through PIAS3-STAT3.
Zhang C, Mukherjee S, Tucker-Burden C, Ross JL, Chau MJ, Kong J, Brat DJ. Zhang C, et al. Mol Oncol. 2017 Mar;11(3):280-294. doi: 10.1002/1878-0261.12034. Epub 2017 Feb 15. Mol Oncol. 2017. PMID: 28100038 Free PMC article. - Stem cell signature in glioblastoma: therapeutic development for a moving target.
Nakano I. Nakano I. J Neurosurg. 2015 Feb;122(2):324-30. doi: 10.3171/2014.9.JNS132253. Epub 2014 Nov 14. J Neurosurg. 2015. PMID: 25397368 Review. - Collateral damage control in cancer therapy: defining the stem identity in gliomas.
Hsieh D. Hsieh D. Curr Pharm Des. 2011;17(23):2370-85. doi: 10.2174/138161211797249198. Curr Pharm Des. 2011. PMID: 21827417 Review.
Cited by
- PI3K and STAT3: a new alliance.
Vogt PK, Hart JR. Vogt PK, et al. Cancer Discov. 2011 Nov;1(6):481-6. doi: 10.1158/2159-8290.CD-11-0218. Cancer Discov. 2011. PMID: 22348200 Free PMC article. Review. - Tetraspanin CD9 stabilizes gp130 by preventing its ubiquitin-dependent lysosomal degradation to promote STAT3 activation in glioma stem cells.
Shi Y, Zhou W, Cheng L, Chen C, Huang Z, Fang X, Wu Q, He Z, Xu S, Lathia JD, Ping Y, Rich JN, Bian XW, Bao S. Shi Y, et al. Cell Death Differ. 2017 Jan;24(1):167-180. doi: 10.1038/cdd.2016.110. Epub 2016 Oct 14. Cell Death Differ. 2017. PMID: 27740621 Free PMC article. - Epidermal growth factor receptor as a therapeutic target in glioblastoma.
Kalman B, Szep E, Garzuly F, Post DE. Kalman B, et al. Neuromolecular Med. 2013 Jun;15(2):420-34. doi: 10.1007/s12017-013-8229-y. Epub 2013 Apr 11. Neuromolecular Med. 2013. PMID: 23575987 Review. - CD90 is identified as a candidate marker for cancer stem cells in primary high-grade gliomas using tissue microarrays.
He J, Liu Y, Zhu T, Zhu J, Dimeco F, Vescovi AL, Heth JA, Muraszko KM, Fan X, Lubman DM. He J, et al. Mol Cell Proteomics. 2012 Jun;11(6):M111.010744. doi: 10.1074/mcp.M111.010744. Epub 2011 Dec 27. Mol Cell Proteomics. 2012. PMID: 22203689 Free PMC article. - The impact of STAT3 and phospho-STAT3 expression on the prognosis and clinicopathology of ovarian cancer: a systematic review and meta-analysis.
Gao S, Zhang W, Yan N, Li M, Mu X, Yin H, Wang J. Gao S, et al. J Ovarian Res. 2021 Nov 18;14(1):164. doi: 10.1186/s13048-021-00918-6. J Ovarian Res. 2021. PMID: 34789292 Free PMC article.
References
- Bao S, Wu Q, McLendon RE, Hao Y, Shi Q, Hjelmeland AB, Dewhirst MW, Bigner DD, Rich JN. GBM stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006a;444:756–760. - PubMed
- Bao S, Wu Q, Sathornsumetee S, Hao Y, Li Z, Hjelmeland AB, Shi Q, McLendon RE, Bigner DD, Rich JN. Stem Cell-like GBM Cells Promote Tumor Angiogenesis through Vascular Endothelial Growth Factor. Cancer Res. 2006b;66:7843–7848. - PubMed
- Beier D, Hau P, Proescholdt M, Lohmeier A, Wischhusen J, Oefner PJ, Aigner L, Brawanski A, Bogdahn U, Beier CP. CD133(+) and CD133(−) glioblastoma-derived cancer stem cells show differential growth characteristics and molecular profiles. Cancer Res. 2007;67:4010–4015. - PubMed
- Bromberg JF, Wrzeszczynska MH, Devgan G, Zhao Y, Pestell RG, Albanese C, Darnell JE., Jr. Stat3 as an oncogene. Cell. 1999;98:295–303. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- NS054276/NS/NINDS NIH HHS/United States
- NS070315/NS/NINDS NIH HHS/United States
- P01 HL085098-04/HL/NHLBI NIH HHS/United States
- R01 NS070315-02/NS/NINDS NIH HHS/United States
- CA129958/CA/NCI NIH HHS/United States
- R01 HL081092/HL/NHLBI NIH HHS/United States
- R01 HL081092-05/HL/NHLBI NIH HHS/United States
- R01 NS070315-01/NS/NINDS NIH HHS/United States
- P01 HL085098/HL/NHLBI NIH HHS/United States
- R01 CA129958/CA/NCI NIH HHS/United States
- R01 NS070315/NS/NINDS NIH HHS/United States
- R01 CA116659/CA/NCI NIH HHS/United States
- CA154130/CA/NCI NIH HHS/United States
- CA116659/CA/NCI NIH HHS/United States
- R01 NS054276/NS/NINDS NIH HHS/United States
- R01 CA154130/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous