A genetically encoded tag for correlated light and electron microscopy of intact cells, tissues, and organisms - PubMed (original) (raw)
A genetically encoded tag for correlated light and electron microscopy of intact cells, tissues, and organisms
Xiaokun Shu et al. PLoS Biol. 2011 Apr.
Abstract
Electron microscopy (EM) achieves the highest spatial resolution in protein localization, but specific protein EM labeling has lacked generally applicable genetically encoded tags for in situ visualization in cells and tissues. Here we introduce "miniSOG" (for mini Singlet Oxygen Generator), a fluorescent flavoprotein engineered from Arabidopsis phototropin 2. MiniSOG contains 106 amino acids, less than half the size of Green Fluorescent Protein. Illumination of miniSOG generates sufficient singlet oxygen to locally catalyze the polymerization of diaminobenzidine into an osmiophilic reaction product resolvable by EM. MiniSOG fusions to many well-characterized proteins localize correctly in mammalian cells, intact nematodes, and rodents, enabling correlated fluorescence and EM from large volumes of tissue after strong aldehyde fixation, without the need for exogenous ligands, probes, or destructive permeabilizing detergents. MiniSOG permits high quality ultrastructural preservation and 3-dimensional protein localization via electron tomography or serial section block face scanning electron microscopy. EM shows that miniSOG-tagged SynCAM1 is presynaptic in cultured cortical neurons, whereas miniSOG-tagged SynCAM2 is postsynaptic in culture and in intact mice. Thus SynCAM1 and SynCAM2 could be heterophilic partners. MiniSOG may do for EM what Green Fluorescent Protein did for fluorescence microscopy.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
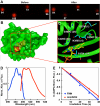
Figure 1. MiniSOG, a small and efficient singlet oxygen generator, is engineered from a blue light photoreceptor based on protein crystal structure.
(A) Infrared fluorescence of E. coli colonies expressing the fusion proteins before and after irradiation (480±15 nm excitation). (B) Predicted structure of miniSOG by the Swiss-Model structure homology-modeling server . (C) Mutations introduced into miniSOG compared to its parent. Numbers in bracket are based on miniSOG protein sequence. (D) Normalized absorbance (blue) and emission (red) spectra. (E) Degradation of ADPA by illumination of miniSOG (red) or free FMN (blue).
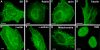
Figure 2. MiniSOG-labeled proteins and organelles exhibit correct localization at the light microscopic level.
Confocal fluorescence images of miniSOG-targeted endoplasmic reticulum (A), Rab5a (B), zyxin (C), tubulin (D), β-actin (E), α-actinin (F), mitochondria (G), and histone 2B (H) in HeLa cells; scale bars, 10 µm.
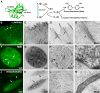
Figure 3. MiniSOG produces correlated fluorescence and EM contrast with correct localization of labeled proteins and organelles.
(A) Schematic diagram of how miniSOG produces EM contrast upon blue-light illumination. Spin states are depicted by the arrows. ISC, intersystem crossing. Correlated confocal fluorescence (B,F,J), transmitted light (C,G,K), and electron microscopic (D,E,H,I,L,M) imaging of a variety of proteins. (B–E) HeLa cells expressing miniSOG labeled α-actinin. Arrows denote correlated structures. (F–I) Histone 2B. Panel H is a 3 nm thick computed slice from an electron tomogram. Panel I is a high magnification thin section electron micrograph showing labeled chromatin fibers near the nuclear envelope (arrows) and a nuclear pore (arrowhead). (J–M) Mitochondrial targeted miniSOG. Panels J and K show a confocal image prior to photooxidation and a transmitted light image following photooxidation, respectively. The differential contrast generated between a transfected (arrows) and non-transfected cell (arrowheads) is evident. Bars B–D, 1 micron; E, 200 nm; F–H, 2 microns; I, 100 nm; J–L, 5 microns; M, 200 nm.
Figure 4. MiniSOG-tagged Cx43 forms gap junctions.
(A) The green fluorescence of miniSOG reveals gap junctions and transporting vesicles. (B) Electron microscopy indicates negatively stained structures of appropriate size and spacing to be gap junction channels (arrows). (C) Studs on the membranes of trafficking vesicles suggest single connexons. The arrowhead points to two dots with a center-to-center distance ∼14 nm. (D) A high-quality immunogold image showing a randomly labeled fraction of densely packed Cx43 gap junctions. This figure is reproduced from Figure 4D of Gaietta et al. . (E) A cartoon showing miniSOG-labeled Cx43 gap junctions. Bar A, 10 microns; B–D, 100 nm.
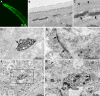
Figure 5. MiniSOG produces fluorescence and EM contrast in C. elegans and reveals previously unknown localization of synaptic cell adhesion molecules in mice.
(A) Confocal fluorescence image of miniSOG targeted to the mitochondria in body wall muscles of C. elegans. (B–C) Thin section EM images of a portion of C. elegans showing a subset of labeled mitochondria in the body wall muscle (arrow) and adjacent unlabeled mitochondria in a different cell type (arrowheads). (D–E) Ultrastructural localization of miniSOG-labeled synaptic cell-adhesion molecules (SynCAMs) in cultured cortical neurons. (D) SynCAM1 fusion reveals uniform membrane labeling at the presynaptic apposition (arrow). (E) SynCAM2 fusion shows postsynaptic membrane labeling (pointed by arrow). Ultrastructural details including synaptic vesicles and nerve terminal substructure were well preserved in both (D) and (E). (F–G) Ultrastructural localization of miniSOG-labeled synaptic cell-adhesion molecule 2 (SynCAM2) in intact mouse brain. (A) A large area (∼14 µm × 14 µm) of one of the tissue sections imaged by serial block-face scanning electron microscopy. (B) Enlargement of the region boxed in (A) reveals postsynaptic membrane labeling (pointed by arrow) apposing a presynaptic bouton containing vesicles. Ultrastructural details including synaptic vesicles and membrane-bound structures of synapses were well preserved and easily recognizable (e.g. arrowhead in the upper left). Bar A, 50 microns; B–C, 500 nm; D–E, 500 nm; F, 2 microns; G, 500 nm.
Similar articles
- The use of miniSOG in the localization of mitochondrial proteins.
Perkins GA. Perkins GA. Methods Enzymol. 2014;547:165-79. doi: 10.1016/B978-0-12-801415-8.00010-2. Methods Enzymol. 2014. PMID: 25416358 - Identification of PSD-95 in the Postsynaptic Density Using MiniSOG and EM Tomography.
Chen X, Winters C, Crocker V, Lazarou M, Sousa AA, Leapman RD, Reese TS. Chen X, et al. Front Neuroanat. 2018 Dec 7;12:107. doi: 10.3389/fnana.2018.00107. eCollection 2018. Front Neuroanat. 2018. PMID: 30581381 Free PMC article. - Photo-inducible cell ablation in Caenorhabditis elegans using the genetically encoded singlet oxygen generating protein miniSOG.
Qi YB, Garren EJ, Shu X, Tsien RY, Jin Y. Qi YB, et al. Proc Natl Acad Sci U S A. 2012 May 8;109(19):7499-504. doi: 10.1073/pnas.1204096109. Epub 2012 Apr 24. Proc Natl Acad Sci U S A. 2012. PMID: 22532663 Free PMC article. - Applications of genetically encoded photosensitizer miniSOG: from correlative light electron microscopy to immunophotosensitizing.
Souslova EA, Mironova KE, Deyev SM. Souslova EA, et al. J Biophotonics. 2017 Mar;10(3):338-352. doi: 10.1002/jbio.201600120. Epub 2016 Jul 20. J Biophotonics. 2017. PMID: 27435584 Review. - Visualizing viral protein structures in cells using genetic probes for correlated light and electron microscopy.
Ou HD, Deerinck TJ, Bushong E, Ellisman MH, O'Shea CC. Ou HD, et al. Methods. 2015 Nov 15;90:39-48. doi: 10.1016/j.ymeth.2015.06.002. Epub 2015 Jun 9. Methods. 2015. PMID: 26066760 Free PMC article. Review.
Cited by
- Promising tools into oxidative stress: A review of non-rodent model organisms.
Zhang Y, Li Y, Ren T, Duan JA, Xiao P. Zhang Y, et al. Redox Biol. 2024 Nov;77:103402. doi: 10.1016/j.redox.2024.103402. Epub 2024 Oct 16. Redox Biol. 2024. PMID: 39437623 Free PMC article. Review. - Neuronal Activity and CaMKII Regulate Kinesin-Mediated Transport of Synaptic AMPARs.
Hoerndli FJ, Wang R, Mellem JE, Kallarackal A, Brockie PJ, Thacker C, Madsen DM, Maricq AV. Hoerndli FJ, et al. Neuron. 2015 Apr 22;86(2):457-74. doi: 10.1016/j.neuron.2015.03.011. Epub 2015 Apr 2. Neuron. 2015. PMID: 25843407 Free PMC article. - A new low cost wide-field illumination method for photooxidation of intracellular fluorescent markers.
da Silva Filho M, Santos DV, Costa KM. da Silva Filho M, et al. PLoS One. 2013;8(2):e56512. doi: 10.1371/journal.pone.0056512. Epub 2013 Feb 18. PLoS One. 2013. PMID: 23441199 Free PMC article. - Natural and Designed Toxins for Precise Therapy: Modern Approaches in Experimental Oncology.
Shilova O, Shramova E, Proshkina G, Deyev S. Shilova O, et al. Int J Mol Sci. 2021 May 7;22(9):4975. doi: 10.3390/ijms22094975. Int J Mol Sci. 2021. PMID: 34067057 Free PMC article. Review. - Corollary discharge promotes a sustained motor state in a neural circuit for navigation.
Ji N, Venkatachalam V, Rodgers HD, Hung W, Kawano T, Clark CM, Lim M, Alkema MJ, Zhen M, Samuel AD. Ji N, et al. Elife. 2021 Apr 21;10:e68848. doi: 10.7554/eLife.68848. Elife. 2021. PMID: 33880993 Free PMC article.
References
- Tsien R. Y. Constructing and exploiting the fluorescent protein paintbox (Nobel Lecture). Angew Chem Int Ed Engl. 2009;48:5612–5626. - PubMed
- Nishino Y, Yasunaga T, Miyazawa A. A genetically encoded metallothionein tag enabling efficient protein detection by electron microscopy. J Electron Microsc (Tokyo) 2007;56(3):93–101. - PubMed
- Diestra E, Fontana J, Guichard P, Marco S, Risco C. Visualization of proteins in intact cells with a clonable tag for electron microscopy. J Struct Biol. 2009;165(3):157–168. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 NS035546/NS/NINDS NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- NS035546/NS/NINDS NIH HHS/United States
- P41 RR004050-24/RR/NCRR NIH HHS/United States
- P41 RR004050/RR/NCRR NIH HHS/United States
- R01 NS027177/NS/NINDS NIH HHS/United States
- P41-RR004050/RR/NCRR NIH HHS/United States
- R37 NS027177/NS/NINDS NIH HHS/United States
- GM086197/GM/NIGMS NIH HHS/United States
- R01 GM086197/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials