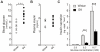Differential glucose-regulation of microRNAs in pancreatic islets of non-obese type 2 diabetes model Goto-Kakizaki rat - PubMed (original) (raw)
Differential glucose-regulation of microRNAs in pancreatic islets of non-obese type 2 diabetes model Goto-Kakizaki rat
Jonathan Lou S Esguerra et al. PLoS One. 2011.
Abstract
Background: The Goto-Kakizaki (GK) rat is a well-studied non-obese spontaneous type 2 diabetes (T2D) animal model characterized by impaired glucose-stimulated insulin secretion (GSIS) in the pancreatic beta cells. MicroRNAs (miRNAs) are short regulatory RNAs involved in many fundamental biological processes. We aim to identify miRNAs that are differentially-expressed in the pancreatic islets of the GK rats and investigate both their short- and long term glucose-dependence during glucose-stimulatory conditions.
Methodology/principal findings: Global profiling of 348 miRNAs in the islets of GK rats and Wistar controls (females, 60 days, N = 6 for both sets) using locked nucleic acid (LNA)-based microarrays allowed for the clear separation of the two groups. Significant analysis of microarrays (SAM) identified 30 differentially-expressed miRNAs, 24 of which are predominantly upregulated in the GK rat islets. Monitoring of qPCR-validated miRNAs during GSIS experiments on isolated islets showed disparate expression trajectories between GK and controls indicating distinct short- and long-term glucose dependence. We specifically found expression of rno-miR-130a, rno-miR-132, rno-miR-212 and rno-miR-335 to be regulated by hyperglycaemia. The putative targets of upregulated miRNAs in the GK, filtered with glucose-regulated mRNAs, were found to be enriched for insulin-secretion genes known to be downregulated in T2D patients. Finally, the binding of rno-miR-335 to a fragment of the 3'UTR of one of known down-regulated exocytotic genes in GK islets, Stxbp1 was shown by luciferase assay.
Conclusions/significance: The perturbed miRNA network found in the GK rat islets is indicative of a system-wide impairment in the regulation of genes important for the normal functions of pancreatic islets, particularly in processes involving insulin secretion during glucose stimulatory conditions. Our findings suggest that the reduced insulin secretion observed in the GK rat may be partly due to upregulated miRNA expression leading to decreased production of key proteins of the insulin exocytotic machinery.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
Figure 1. Phenotype of the GK rats at the time of islets collection.
A. Non-fasting intra-cardial blood glucose levels are elevated in the GK rats compared to non-diabetic Wistar controls (N = 10 in both groups). B. Plasma insulin levels are at comparable levels between the two groups of animals (N = 8 in both groups). C. Insulin secretion is reduced in the isolated pancreatic islets of GK rat at 8.3 mM and 16.7 mM glucose (N = 3 independent RIA in quadruplicate per assay). Data are average ± SEM; (***) P<0.001 GK vs Wistar.
Figure 2. Global miRNA profiles of the pancreatic islets of GK and Wistar rats and qPCR validation.
A. Hierarchical clustering of array signals from 348 rat miRNAs allowed for the separation of the individual animals into two groups, 6 GK vs 6 Wistar. Rat miR-375, miR-132 and miR-708 are indicated for reference, representing no significant change (dark tones), upregulated (yellow tones) and downregulated (blue tones) miRNAs in GK. B. Significant Analysis of Microarrays (SAM) identified 30 differentially-regulated miRNAs in the GK rat pancreatic islets, clustered into 6 downregulated and 24 upregulated miRNAs (median False Discovery Rate = 0%). C. Stem-loop qPCR validation of selected 12 miRNAs in GK and Wistar islets. Mir-375 was included as a non-regulated control. Each miRNA was normalized to the geometric mean of U6 snRNA and U87 rat expressions as implemented in GeNorm v3.5. The 2-ΔΔCt method was used for relative quantification using the Wistar expression level as calibrator. The presented data are the average of N = 3 biological replicates performed independently each in triplicate qPCR wells ± SEM; (*) P<0.05, (**) P<0.01, (***) P<0.001 GK vs Wistar.
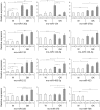
Figure 3. Glucose-dependence of miRNA expression after 1 h incubation at 2.8 mM, 8.3 mM and 16.7 mM glucose.
More pronounced variation of miRNA expression trajectories (magnitude and direction of expression) were observed in the GK compared to Wistar islets across the different glucose concentrations. Each miRNA was normalized to the geometric mean of U6 snRNA and U87 rat. The 2-ΔΔCt method was used for relative quantification using Wistar expression level at 2.8G as calibrator. The presented data are the average of N = 3 biological replicates performed independently each in triplicate qPCR wells ± SEM. Intra-sample (within same animal group) significance denoted by (*) P<0.05, (**) P<0.01, (***) P<0.001 vs 2.8G of same animal group. Inter-sample (W vs GK) significance denoted by (†) P<0.05, (††) P<0.01, (†††) P<0.001, compares expression levels from different animal groups of the same incubating glucose concentration. Different y-axis scaling was used for each miRNA to allow easy comparison of expression levels across different conditions.
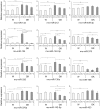
Figure 4. Glucose-dependence of miRNA levels after 24 h incubation at 2.8 mM, 8.3 mM and 16.7 mM glucose.
Three general trends of miRNA expression trajectories were observed for Wistar islets at 2.8G vs 16.7G: i) increased expression as exhibited by rno-miR-132, rno-miR-212 and rno-miR-409-3p, ii) decreased expression as in the case of rno-miR-124, rno-miR-142-3p, rno-miR-375, rno-miR-335, rno-miR-130a and rno-miR-708 and, iii) no change as seen in rno-miR-376a, rno-miR-142-5p and rno-miR-433. For GK islet expression, the miRNAs generally exhibited expression trajectories aimed at attaining Wistar islet levels. Data analysis and presentation are as described for Figure 3.
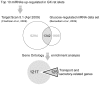
Figure 5. In silico strategy to analyze the miRNAs upregulated in the GK rat islets.
One of the highlights of the approach is the filtering of predicted targets with known glucose-regulated mRNA data set from a previous study, significantly reducing false positive targets. This also resulted to a more focused enrichment of genes already implicated in islet functions.
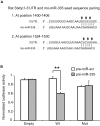
Figure 6. Interaction of rno-miR-335 with the predicted binding sites in the 3′UTR of rat Stxbp1.
A. The rat Stxbp1 3′UTR contains two putative miR-335 binding sites as predicted by TargetScan 5.1. The two sites are located in the proximal region and both could be included in a 200 bp fragment cloned into a dual luciferase reporter plasmid. Arrows indicate the sites in the seed sequences that were mutated into complementary bases to act as negative control in the luciferase assay B. Two inserts, one with the wildtype rat Stxbp1-3′UTR sequence (Wt) and the other with sequences mutated at both miR-335 predicted binding sites (Mut) were cloned into the pmiRGLO dual luciferase vector. The pmiRGLO dual luciferase vector alone (Empty) was included as positive controls. HeLa cells were co-transfected with the empty vector or plasmid constructs and pre-miR-335 or pre-miR-scr (control with scrambled sequence) and assayed after 48 hours. Transfection efficiency was normalized using the Renilla signal. Data represents two independent transfections ± S.E.M. with n = 3. (**) P<0.01.
Similar articles
- Global expression profiling of glucose-regulated genes in pancreatic islets of spontaneously diabetic Goto-Kakizaki rats.
Ghanaat-Pour H, Huang Z, Lehtihet M, Sjöholm A. Ghanaat-Pour H, et al. J Mol Endocrinol. 2007 Aug;39(2):135-50. doi: 10.1677/JME-07-0002. J Mol Endocrinol. 2007. PMID: 17693612 - Overexpression of mitochondrial FAD-linked glycerol-3-phosphate dehydrogenase does not correct glucose-stimulated insulin secretion from diabetic GK rat pancreatic islets.
Ueda K, Tanizawa Y, Ishihara H, Kizuki N, Ohta Y, Matsutani A, Oka Y. Ueda K, et al. Diabetologia. 1998 Jun;41(6):649-53. doi: 10.1007/s001250050963. Diabetologia. 1998. PMID: 9662045 - Inhibition of protein-tyrosine phosphatases stimulates insulin secretion in pancreatic islets of diabetic Goto-Kakizaki rats.
Chen J, Ostenson CG. Chen J, et al. Pancreas. 2005 May;30(4):314-7. doi: 10.1097/01.mpa.0000161887.25115.6c. Pancreas. 2005. PMID: 15841039 - Islet gene expression and function in type 2 diabetes; studies in the Goto-Kakizaki rat and humans.
Ostenson CG, Efendic S. Ostenson CG, et al. Diabetes Obes Metab. 2007 Nov;9 Suppl 2:180-6. doi: 10.1111/j.1463-1326.2007.00787.x. Diabetes Obes Metab. 2007. PMID: 17919192 Review. - miRNAs in the Beta Cell-Friends or Foes?
Karagiannopoulos A, Cowan E, Eliasson L. Karagiannopoulos A, et al. Endocrinology. 2023 Mar 13;164(5):bqad040. doi: 10.1210/endocr/bqad040. Endocrinology. 2023. PMID: 36869830 Free PMC article. Review.
Cited by
- Induction of miR-132 and miR-212 Expression by Glucagon-Like Peptide 1 (GLP-1) in Rodent and Human Pancreatic β-Cells.
Shang J, Li J, Keller MP, Hohmeier HE, Wang Y, Feng Y, Zhou HH, Shen X, Rabaglia M, Soni M, Attie AD, Newgard CB, Thornberry NA, Howard AD, Zhou YP. Shang J, et al. Mol Endocrinol. 2015 Sep;29(9):1243-53. doi: 10.1210/me.2014-1335. Epub 2015 Jul 28. Mol Endocrinol. 2015. PMID: 26218441 Free PMC article. - MicroRNAs in pancreatic cancer metabolism.
Singh PK, Brand RE, Mehla K. Singh PK, et al. Nat Rev Gastroenterol Hepatol. 2012 Apr 17;9(6):334-44. doi: 10.1038/nrgastro.2012.63. Nat Rev Gastroenterol Hepatol. 2012. PMID: 22508159 Free PMC article. Review. - Prospecting of exosomal-miRNA signatures as prognostic marker for gestational diabetes mellitus and other adverse pregnancy outcomes.
Mitra T, Gulati R, Uppal A, Kumari SR, Tripathy S, Ranjan P, Janardhanan R. Mitra T, et al. Front Endocrinol (Lausanne). 2023 Feb 9;14:1097337. doi: 10.3389/fendo.2023.1097337. eCollection 2023. Front Endocrinol (Lausanne). 2023. PMID: 36843574 Free PMC article. Review. - Differential expression and release of exosomal miRNAs by human islets under inflammatory and hypoxic stress.
Saravanan PB, Vasu S, Yoshimatsu G, Darden CM, Wang X, Gu J, Lawrence MC, Naziruddin B. Saravanan PB, et al. Diabetologia. 2019 Oct;62(10):1901-1914. doi: 10.1007/s00125-019-4950-x. Epub 2019 Aug 1. Diabetologia. 2019. PMID: 31372667 - The Profiling and Role of miRNAs in Diabetes Mellitus.
Kim M, Zhang X. Kim M, et al. J Diabetes Clin Res. 2019;1(1):5-23. doi: 10.33696/diabetes.1.003. J Diabetes Clin Res. 2019. PMID: 32432227 Free PMC article.
References
- Winter J, Jung S, Keller S, Gregory RI, Diederichs S. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat Cell Biol. 2009;11:228–234. - PubMed
- Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can up-regulate translation. Science. 2007;318:1931–1934. - PubMed
- Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, et al. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. - PubMed
- Aravin A, Tuschl T. Identification and characterization of small RNAs involved in RNA silencing. FEBS Lett. 2005;579:5830–5840. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical