Mediator head subcomplex Med11/22 contains a common helix bundle building block with a specific function in transcription initiation complex stabilization - PubMed (original) (raw)
. 2011 Aug;39(14):6291-304.
doi: 10.1093/nar/gkr229. Epub 2011 Apr 15.
Affiliations
- PMID: 21498544
- PMCID: PMC3152362
- DOI: 10.1093/nar/gkr229
Mediator head subcomplex Med11/22 contains a common helix bundle building block with a specific function in transcription initiation complex stabilization
Martin Seizl et al. Nucleic Acids Res. 2011 Aug.
Abstract
Mediator is a multiprotein co-activator of RNA polymerase (Pol) II transcription. Mediator contains a conserved core that comprises the 'head' and 'middle' modules. We present here a structure-function analysis of the essential Med11/22 heterodimer, a part of the head module. Med11/22 forms a conserved four-helix bundle domain with C-terminal extensions, which bind the central head subunit Med17. A highly conserved patch on the bundle surface is required for stable transcription pre-initiation complex formation on a Pol II promoter in vitro and in vivo and may recruit the general transcription factor TFIIH. The bundle domain fold is also present in the Mediator middle module subcomplex Med7/21 and is predicted in the Mediator heterodimers Med2/3, Med4/9, Med10/14 and Med28/30. The bundle domain thus represents a common building block that has been multiplied and functionally diversified during Mediator evolution in eukaryotes.
Figures
Figure 1.
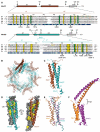
Structure of Med11/22 Mediator subcomplex. (A) Multiple sequence alignment of the conserved core of Med11 and Med22 from Saccharomyces cerevisiae (Sc), Schizosaccharomyces pombe (Sp), Caenorhabditis elegans (Ce), Drosophila melanogaster (Dm) and Homo sapiens (Hs). The corrected numbering of Sc Med11 is used. Invariant and conserved residues are highlighted in green and yellow, respectively. Additionally, residues that are invariant or conserved among the yeast family Saccharomycotinae (Sc, Candida glabrata, Candida albicans, Ashbya gossypii, Kluyveromyce lactis and Debaryomyces hansenii) are highlighted with green and yellow frames on the Sc sequence, respectively. Surface accessibility is indicated below the sequences (blue, high; cyan, intermediate; white, buried). Secondary structure elements of the conserved structural core are shown above the sequences (spirals, α-helices; lines, ordered but without secondary structure; dashed lines, disordered in crystal construct). In addition schematic views of the full proteins are shown. Consensus secondary structure predictions (61–63) for the C-termini of Sc Med11 and Med22 are indicated with dashed lines and gray filling. All truncations relevant for this study are indicated with arrows and the residue number. Previously reported as well as mutations generated in this study are marked with filled triangles. Sequence alignments were done with MUSCLE (64) and figures were prepared with ESPript (65). (B) Ribbon-model representation of the 12 heterodimers within the asymmetric unit. Med11 and Med22 are depicted in brown and cyan, respectively. (C) Ribbon-model representation of the Med11/22 crystal structure. Secondary structure elements are labeled according to (A). The linker between helices α1 and α* of Med22 (residues 33–40) was disordered. C-termini of Med11 and Med22 had to be removed from crystallization construct and are therefore lacking in the structure. (D) Dimer interface conservation. On the left, Med11 is shown in surface representation together with a ribbon model of Med22. The view is related to (C) by a 90° rotation around a vertical axis. On the right, Med22 is shown in surface representation together with a ribbon model of Med11. The two views are related by a 180° rotation around a vertical axis. (E) Superimposition of Cα traces of the four-helix bundle folds of Med11/22 (brown and cyan; this study) and Med7C/21 [orange and purple; (31)]. (F) Ribbon-model representation of the complete Med7C/21 structure (pdb accession code 1YKE). The reported flexible hinge region is indicated.
Figure 2.
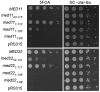
C-terminal extensions of Med11/22 are essential for viability. Yeast complementation assays. MED11 and MED22 constructs including 500 bp upstream of the start codon and 300 bp downstream of the stop codon were cloned into a pRS315 plasmid (LEU2) and transformed into the respective yeast shuffle strains. On 5FOA plates the URA3 shuffle plasmid encoding the respective full-length gene is shuffled out. Yeast cells lacking either the N- or C-terminus of Med11 or the C-terminus of Med22 are inviable. Cells lacking the N-terminal helix of Med22 or the last 10 amino acids of Med11 display a slow growth phenotype.
Figure 3.
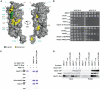
A heterodimeric four-helix bundle building block in Mediator. Schematic depiction of Mediator subunits predicted to share the heterodimeric bundle fold. α-helices from crystal structures or from predictions (61–63) are indicated as boxes drawn to scale. Helix α1, α2 and C-terminal helical extensions are colored in black, gray and white, respectively. Check marks indicate experimentally confirmed heterodimers (co-expression and co-purification). Structural homology to the published Med7C/21 structure was predicted with HHPred (
http://toolkit.lmb.uni-muenchen.de/hhpred
) for all listed subunits except the highly divergent Sc subunits Med2 and Med3. The _P_-value and score for the HHPred searches are given. When the human protein sequence was used for the HHPred search, the _P_-values are marked with an asterisk.
Figure 4.
C-terminal extensions of Med11/22 bind a Med17 C-terminal domain. (A) Co-expression in E. coli and co-purification of Sc a C-terminal domain of Med17C (residues 377–687) with Sc 6His-Med22/Med11 constructs using nickel magnetic beads. A co-purifying contaminant is marked with an asterisk. (B) Co-immunoprecipitation of the putative Sp Med11-3HA with Sp Med7-TAP from Sp lysate using IgG agarose beads. Med7 was eluted natively using tobacco etch virus protease. Med7 was detected in the input using peroxidase/anti-peroxidase antibody complex. Med11 was detected in both input and eluate using anti-HA antibody. (C) Co-expression in E. coli and co-purification of Sp Med17 (lanes 1–3) and Sp Med17C (residues 257–545; lanes 4–6) with Sp His6-Med22/Med11 constructs using nickel magnetic beads.
Figure 5.
A conserved interaction patch on Med11/22. (A) Surface conservation of the Med11/22 four-helix bundle. Invariant and conserved residues from yeast to human are highlighted in green and yellow, respectively. Residues of Med11 (brown) and Med22 (cyan) targeted by mutagenesis in this study and in previous reports are indicated with arrows. The orientation is identical to Figure 1C. The two views are related by a 180° rotation around the vertical axis. (B) Spot dilutions of yeast strains carrying structure-based mutations on the conserved surface patch and viable truncations (Figure 2) on YPD plates at 30°C and 37°C. (C) Co-expression in E. coli and co-purification of Sc Med17C with Sc 6His-Med22/Med11 and Sc 6His-Med22/Med11-E17K/L24K using nickel magnetic beads. (D) Co-immunoprecipitation of Sc Med11-3HA (Mediator head module) and Sc Med2 (Mediator tail module) with Sc Med7-TAP (Mediator middle module) from wild-type, _med11_1–105 and med11-E17K/L24K yeast strains.
Figure 6.
Med11/22 is a functional Mediator submodule in vitro and in vivo. (A) Hierarchical cluster diagram (Pearson correlation) of genes exhibiting significantly altered mRNA levels (>2-fold, vertical axis) for different Mediator mutant strains (horizontal axis). Changes in mRNA levels compared to the wild-type strain are depicted in yellow (up), blue (down) or black (no change). (B) Pearson correlation matrix for expression profiles of different Mediator mutants (1, very high correlation; 0, no correlation; −1, very high anti-correlation). (C) Chromatin immunoprecipitation of Rpb3-TAP (Pol II) and Kin28-TAP (TFIIH kinase module) in wild-type, med11_1–105 and med11-E17K/L24K strains grown to early exponential phase in YPD medium containing 2% glucose. Fold enrichment over a heterochromatic control region is shown for the yeast promoters of a highly expressed gene (ILV5), a housekeeping gene (ADH1) and a glucose-repressed gene (GAL1). (D) In vitro PIC assembly of wild-type, med11_1–105 and med11-E17K/L24K nuclear extracts (NE) on the immobilized HIS4 yeast promoter. PIC formation of mutant extracts was partially rescued by adding 2 pmol tandem-affinity purified Mediator complex (TAP-Med) to the nuclear extracts prior to PIC assembly. Presence of Pol II (Rpb3 & Rpb11), TFIIB, Mediator (Med17) and TFIID (Taf4) was tested by western Blot. (E) In vitro transcription assay with wild-type, _med11_1–105 and med11-E17K/L24K nuclear extracts (NE) on the HIS4 yeast promoter. Order of addition is shown on top. Transcription was partially rescued by adding 2 pmol tandem-affinity purified Mediator complex (TAP-Med).
Figure 7.
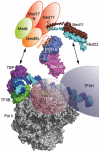
Submodular architecture of the Mediator head and relative size of the Pol II PIC. Crystal structures of the essential Med11/22 four-helix bundle (this article), the previously described non-essential Med8C/18/20 subcomplex (29) and a molecular model of the Pol II–TBP–TFIIB–DNA promoter closed complex (66) are drawn to scale. Locations of the general factors TFIIF and H, as determined by biochemical probing (67,68), are indicated by semi-transparent ellipsoids. Mediator head interactions with PIC components, namely Med8/18/20-TBP (19) and Med11/22–TFIIH (20), are indicated by arrows.
Similar articles
- Structure of the Mediator head module.
Larivière L, Plaschka C, Seizl M, Wenzeck L, Kurth F, Cramer P. Larivière L, et al. Nature. 2012 Dec 20;492(7429):448-51. doi: 10.1038/nature11670. Epub 2012 Oct 31. Nature. 2012. PMID: 23123849 - Core Mediator structure at 3.4 Å extends model of transcription initiation complex.
Nozawa K, Schneider TR, Cramer P. Nozawa K, et al. Nature. 2017 May 11;545(7653):248-251. doi: 10.1038/nature22328. Epub 2017 May 3. Nature. 2017. PMID: 28467824 - Model of the Mediator middle module based on protein cross-linking.
Larivière L, Plaschka C, Seizl M, Petrotchenko EV, Wenzeck L, Borchers CH, Cramer P. Larivière L, et al. Nucleic Acids Res. 2013 Nov;41(20):9266-73. doi: 10.1093/nar/gkt704. Epub 2013 Aug 11. Nucleic Acids Res. 2013. PMID: 23939621 Free PMC article. - Origins and activity of the Mediator complex.
Conaway RC, Conaway JW. Conaway RC, et al. Semin Cell Dev Biol. 2011 Sep;22(7):729-34. doi: 10.1016/j.semcdb.2011.07.021. Epub 2011 Jul 28. Semin Cell Dev Biol. 2011. PMID: 21821140 Free PMC article. Review. - Twenty years of Mediator complex structural studies.
Verger A, Monté D, Villeret V. Verger A, et al. Biochem Soc Trans. 2019 Feb 28;47(1):399-410. doi: 10.1042/BST20180608. Epub 2019 Feb 7. Biochem Soc Trans. 2019. PMID: 30733343 Free PMC article. Review.
Cited by
- Structure of the Mediator head module.
Larivière L, Plaschka C, Seizl M, Wenzeck L, Kurth F, Cramer P. Larivière L, et al. Nature. 2012 Dec 20;492(7429):448-51. doi: 10.1038/nature11670. Epub 2012 Oct 31. Nature. 2012. PMID: 23123849 - Structure and function of the initially transcribing RNA polymerase II-TFIIB complex.
Sainsbury S, Niesser J, Cramer P. Sainsbury S, et al. Nature. 2013 Jan 17;493(7432):437-40. doi: 10.1038/nature11715. Epub 2012 Nov 14. Nature. 2013. PMID: 23151482 - BRF1 mutations alter RNA polymerase III-dependent transcription and cause neurodevelopmental anomalies.
Borck G, Hög F, Dentici ML, Tan PL, Sowada N, Medeira A, Gueneau L, Thiele H, Kousi M, Lepri F, Wenzeck L, Blumenthal I, Radicioni A, Schwarzenberg TL, Mandriani B, Fischetto R, Morris-Rosendahl DJ, Altmüller J, Reymond A, Nürnberg P, Merla G, Dallapiccola B, Katsanis N, Cramer P, Kubisch C. Borck G, et al. Genome Res. 2015 Feb;25(2):155-66. doi: 10.1101/gr.176925.114. Epub 2015 Jan 5. Genome Res. 2015. PMID: 25561519 Free PMC article. - Mediator recruitment to heat shock genes requires dual Hsf1 activation domains and mediator tail subunits Med15 and Med16.
Kim S, Gross DS. Kim S, et al. J Biol Chem. 2013 Apr 26;288(17):12197-213. doi: 10.1074/jbc.M112.449553. Epub 2013 Feb 27. J Biol Chem. 2013. PMID: 23447536 Free PMC article. - Mechanisms of Mediator complex action in transcriptional activation.
Ansari SA, Morse RH. Ansari SA, et al. Cell Mol Life Sci. 2013 Aug;70(15):2743-56. doi: 10.1007/s00018-013-1265-9. Epub 2013 Jan 30. Cell Mol Life Sci. 2013. PMID: 23361037 Free PMC article. Review.
References
- Roeder RG. The role of general initiation factors in transcription by RNA polymerase II. Trends Biochem. Sci. 1996;21:327–335. - PubMed
- Bjorklund S, Gustafsson C. The yeast Mediator complex and its regulation. Trends Biochem. Sci. 2005;30:240–244. - PubMed
- Kornberg R. Mediator and the mechanism of transcriptional activation. Trends Biochem. Sci. 2005;30:235–239. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials