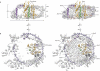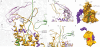Cryo-EM structure of the ribosome-SecYE complex in the membrane environment - PubMed (original) (raw)
Cryo-EM structure of the ribosome-SecYE complex in the membrane environment
Jens Frauenfeld et al. Nat Struct Mol Biol. 2011 May.
Abstract
The ubiquitous SecY-Sec61 complex translocates nascent secretory proteins across cellular membranes and integrates membrane proteins into lipid bilayers. Several structures of mostly detergent-solubilized Sec complexes have been reported. Here we present a single-particle cryo-EM structure of the SecYEG complex in a membrane environment, bound to a translating ribosome, at subnanometer resolution. Using the SecYEG complex reconstituted in a so-called Nanodisc, we could trace the nascent polypeptide chain from the peptidyltransferase center into the membrane. The reconstruction allowed for the identification of ribosome-lipid interactions. The rRNA helix 59 (H59) directly contacts the lipid surface and appears to modulate the membrane in immediate vicinity to the proposed lateral gate of the protein-conducting channel (PCC). On the basis of our map and molecular dynamics simulations, we present a model of a signal anchor-gated PCC in the membrane.
Figures
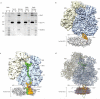
Figure 1. Reconstitution and cryo–EM reconstruction of a 70S RNC–Nd–SecYEG complex
(a) Binding assay using purified RNCs (RNC) with an excess of reconstituted Nd–E and Nd–SecYEG. Supernatant (S) and pellet (P) fractions were analyzed by SDS– polyacrylamide gel electrophoresis and SYPRO orange staining. Nd–SecYEG bind stably to RNCs, whereas Nd–E do not. (b) Cryo–EM reconstruction of the active 70S–RNC–Nd–SecYEG complex at 7.1 Å resolution. The ribosomal 30S subunit is shown in yellow, the 50S subunit blue, SecY orange, SecE purple, Nanodisc white. (c) Density as in (b), but cut perpendicularly to the plane of the membrane along the polypeptide exit tunnel, colours as in b with P–site tRNA and nascent polypeptide chain green. (d) All–atom model of the active 70S–RNC–Nd–SecYEG complex. View and colours as in (b), proteins and RNA in ribbon representation and phospholipids in ball and stick representation with phospholipid headgroups in red/orange and acyl chains white, Apo–A1 in light purple.
Figure 2. Structure of the Nanodisc
(a) Left: side view cut perpendicular to the plane of the membrane of the isolated electron density of the Nanodisc–SecYEG complex (Nd–SecYEG), showing the lateral gate of SecY. The electron density is represented as a transparent grey mesh with the ribbon representation of the fitted model of a SecY (orange), SecE (purple) and the signal anchor sequence (green). Two layers of density are visible (upper membrane interface, UMI, and lower membrane interface, LMI), separated by a region of lower density (hydrophobic core, HC), containing transmembrane (TM) helices. Dimensions are indicated. Right: Same view, with the fitted Nanodisc–model containing lipids in ball and stick representation. Phospholipid headgroups are red (oxygen) and orange (phosphate), acyl chains white (AC, carbon–hydrogen groups). (b) Left: horizontal section, sliced within the plane of the membrane within the hydrophobic core of the lipid bilayer. Rod–like features are visible in the interior of the Nanodisc and account for density of a monomeric SecYEG complex. Right: horizontal section with fitted lipids.
Figure 3. Structure and connections of the membrane–embedded open SecYEG–RNC complex
(a) Cryo–EM reconstruction of the 70S–RNC–Nd–SecYEG complex, colours as in Fig. 1. Insert: molecular model of the 50S subunit with electron density (left) and molecular model for SecYE (right). (b) Model of the open SecYE complex with a signal anchor (SA, green) residing within the lateral gate, view cut perpendicular to the plane of the membrane. (c) Close–up of the interaction area of universal ribosomal adaptor site and SecYE. (d) Molecular model of the ribosome–SecYE–membrane interface with transparent density filtered at 9–10 Å. Lipid headgroups (LH) are indicated. (e) Cytoplasmic view of the molecular model of the Nd–SecYE complex with contacts to the 50S subunit indicated by circles. (f) View of the ribosomal tunnel exit site, contact sites as in (e).
Figure 4. Path of the nascent chain and signal anchor
(a) Section through molecular models of the ribosomal exit tunnel and Nd–SecYE. The nascent chain (NC) with the signal anchor (SA) is shown in green. The dotted line indicates the cytoplasm–membrane interface. (b) Conformational changes of L23. Comparison of the model of L23 (grey) of an inactive ribosome (PDB: 2i2v) and of L23 (pink) in the presence of a nascent chain (green), SecY (orange), SecE (purple) and lipid headgroups. The intra–tunnel loop of L23 bends towards the nascent chain, close to L6/7 of SecY. (c) Conformational change of the β–hairpin loop of L24. Comparison of the model of L24 (grey) of an inactive ribosome (PDB: 2i2v) and of L24 (green) in the presence of a nascent chain (green), SecY (orange), SecY C–terminus (SecY C–term), SecE (purple) and lipid headgroups (LH). (d) View of the lateral gate of SecYE shown as a surface representation. SecY is shown in orange, SecE in purple, the nascent chain in green. Conserved residues of SecY that contribute to the central hydrophobic pore ring are indicated in red and hydrophobic residues of the hydrophobic crevasse that have been found by mutational analysis to be critical for SecY function are indicated in pale yellow. (e) View of the position of the SA from the cytoplasmic side, sliced within the plane of the membrane. Hydrophobic pore ring residues are indicated in red.
Figure 5. Molecular dynamics simulation and membrane insertion
(a) Plot of surface area formed between lipids and ribosomal helix H59 during the MD simulation. (b) Surface representation of the Nd–SecYE complex seen from the ribosome after 16 ns MD simulation. Apo–A1 is shown in light purple, SecY in orange, SecE in purple, nascent chain in green and the atoms of the lipid head–groups are coloured in orange (phosphate), red (oxygen) and blue (nitrogen), respectively. Note the accumulation of positive charges in the region close to H59 and the disorder of the lipids forming a groove juxtaposed to the signal anchor. (c) Schematic depiction of the view in (b) using the same colour code and indicating the probable path of the nascent TM domain for integration into the bilayer. (d) Schematic depiction of the bacterial 50S ribosomal subunit (blue) bound to SRP (red, 4.5 S RNA and the N–terminal 54–NG domain) in the presence of a signal anchor sequence as observed before. The nascent chain with the signal anchor is shown in green and (e) Schematic depiction of a hypothetical TM domain insertion intermediate showing the bacterial 50S ribosomal subunit (blue) bound to the SecYEG complex (orange) in the presence of a signal anchor, accessing the hydrophobic lipid phase through a partially open lateral gate. (f) Schematic depiction of the observed insertion intermediate with the signal anchor TM domain fully inserted into the lateral gate and exposed to the hydrophobic core of the bilayer. Note the proximity of the SA position as observed in the targeting complex (d) and in the insertion intermediate (e, f).
Similar articles
- Structure of the E. coli protein-conducting channel bound to a translating ribosome.
Mitra K, Schaffitzel C, Shaikh T, Tama F, Jenni S, Brooks CL 3rd, Ban N, Frank J. Mitra K, et al. Nature. 2005 Nov 17;438(7066):318-24. doi: 10.1038/nature04133. Nature. 2005. PMID: 16292303 Free PMC article. - Structures of the Sec61 complex engaged in nascent peptide translocation or membrane insertion.
Gogala M, Becker T, Beatrix B, Armache JP, Barrio-Garcia C, Berninghausen O, Beckmann R. Gogala M, et al. Nature. 2014 Feb 6;506(7486):107-10. doi: 10.1038/nature12950. Nature. 2014. PMID: 24499919 - Structure of the SecY channel during initiation of protein translocation.
Park E, Ménétret JF, Gumbart JC, Ludtke SJ, Li W, Whynot A, Rapoport TA, Akey CW. Park E, et al. Nature. 2014 Feb 6;506(7486):102-6. doi: 10.1038/nature12720. Epub 2013 Oct 23. Nature. 2014. PMID: 24153188 Free PMC article. - Translocation of proteins through the Sec61 and SecYEG channels.
Mandon EC, Trueman SF, Gilmore R. Mandon EC, et al. Curr Opin Cell Biol. 2009 Aug;21(4):501-7. doi: 10.1016/j.ceb.2009.04.010. Epub 2009 May 18. Curr Opin Cell Biol. 2009. PMID: 19450960 Free PMC article. Review. - Protein traffic in bacteria: multiple routes from the ribosome to and across the membrane.
Müller M, Koch HG, Beck K, Schäfer U. Müller M, et al. Prog Nucleic Acid Res Mol Biol. 2001;66:107-57. doi: 10.1016/s0079-6603(00)66028-2. Prog Nucleic Acid Res Mol Biol. 2001. PMID: 11051763 Review.
Cited by
- Peptide Folding in Translocon-Like Pores.
Ulmschneider MB, Koehler Leman J, Fennell H, Beckstein O. Ulmschneider MB, et al. J Membr Biol. 2015 Jun;248(3):407-17. doi: 10.1007/s00232-015-9808-7. Epub 2015 May 28. J Membr Biol. 2015. PMID: 26016471 - Parallel Generalized Born Implicit Solvent Calculations with NAMD.
Tanner DE, Chan KY, Phillips JC, Schulten K. Tanner DE, et al. J Chem Theory Comput. 2011 Nov 8;7(11):3635-3642. doi: 10.1021/ct200563j. J Chem Theory Comput. 2011. PMID: 22121340 Free PMC article. - Structure and Dynamics of GPCRs in Lipid Membranes: Physical Principles and Experimental Approaches.
Jones AJY, Gabriel F, Tandale A, Nietlispach D. Jones AJY, et al. Molecules. 2020 Oct 15;25(20):4729. doi: 10.3390/molecules25204729. Molecules. 2020. PMID: 33076366 Free PMC article. Review. - Protein translocation: what's the problem?
Corey RA, Allen WJ, Collinson I. Corey RA, et al. Biochem Soc Trans. 2016 Jun 15;44(3):753-9. doi: 10.1042/BST20160047. Biochem Soc Trans. 2016. PMID: 27284038 Free PMC article. Review. - Analysis of Sec61p and Ssh1p interactions in the ER membrane using the split-ubiquitin system.
Harty C, Römisch K. Harty C, et al. BMC Cell Biol. 2013 Mar 11;14:14. doi: 10.1186/1471-2121-14-14. BMC Cell Biol. 2013. PMID: 23497013 Free PMC article.
References
- Rapoport TA. Protein translocation across the eukaryotic endoplasmic reticulum and bacterial plasma membranes. Nature. 2007;450(7170):663–669. - PubMed
- Halic M, Beckmann R. The signal recognition particle and its interactions during protein targeting. Curr Opin Struct Biol. 2005;15(1):116–125. - PubMed
- Van den Berg B, et al. X–ray structure of a protein–conducting channel. Nature. 2004;427(6969):36–44. Epub 2003 Dec 2003. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P41 RR005969/RR/NCRR NIH HHS/United States
- R01 GM067887/GM/NIGMS NIH HHS/United States
- R01-GM067887/GM/NIGMS NIH HHS/United States
- P41-RR005969/RR/NCRR NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases