Differential microRNA regulation of HLA-C expression and its association with HIV control - PubMed (original) (raw)
. 2011 Apr 28;472(7344):495-8.
doi: 10.1038/nature09914. Epub 2011 Apr 17.
Ram Savan, Ying Qi, Xiaojiang Gao, Yuko Yuki, Sara E Bass, Maureen P Martin, Peter Hunt, Steven G Deeks, Amalio Telenti, Florencia Pereyra, David Goldstein, Steven Wolinsky, Bruce Walker, Howard A Young, Mary Carrington
Affiliations
- PMID: 21499264
- PMCID: PMC3084326
- DOI: 10.1038/nature09914
Differential microRNA regulation of HLA-C expression and its association with HIV control
Smita Kulkarni et al. Nature. 2011.
Abstract
The HLA-C locus is distinct relative to the other classical HLA class I loci in that it has relatively limited polymorphism, lower expression on the cell surface, and more extensive ligand-receptor interactions with killer-cell immunoglobulin-like receptors. A single nucleotide polymorphism (SNP) 35 kb upstream of HLA-C (rs9264942; termed -35) associates with control of HIV, and with levels of HLA-C messenger RNA transcripts and cell-surface expression, but the mechanism underlying its varied expression is unknown. We proposed that the -35 SNP is not the causal variant for differential HLA-C expression, but rather is marking another polymorphism that directly affects levels of HLA-C. Here we show that variation within the 3' untranslated region (UTR) of HLA-C regulates binding of the microRNA hsa-miR-148 to its target site, resulting in relatively low surface expression of alleles that bind this microRNA and high expression of HLA-C alleles that escape post-transcriptional regulation. The 3' UTR variant associates strongly with control of HIV, potentially adding to the effects of genetic variation encoding the peptide-binding region of the HLA class I loci. Variation in HLA-C expression adds another layer of diversity to this highly polymorphic locus that must be considered when deciphering the function of these molecules in health and disease.
Conflict of interest statement
The authors declare no competing financial interests.
Figures
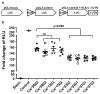
Figure 1. Variation in the HLA-C 3′UTR differentially affects the expression of a reporter gene
Full length 3′UTRs of various HLA-C alleles cloned into luciferase reporter constructs were transfected into B721.221 cell lines and the stability of the mRNA was estimated by dual luciferase reporter assays. The normalized luciferase activity is presented as fold change of relative light units (RLU). The data represent six replicates in each experimental group, the Mean ±SE are depicted as horizontal and vertical bars for each group, respectively, and one of three comparable experiments performed is shown. Non-parametric Wilcoxon-Mann-Whitney tests were used for statistical comparisons and two tailed p values are indicated. ns = not significant. a. Schematic representations of the luciferase reporter constructs used in this study. b. Fold change in luciferase activity of 3′UTRs of HLA-C alleles as compared to that of Cw*0602.
Figure 2. Disruption of miR-148a target site rescues suppression
a. Partial sequence of mutated 3′UTRs of Cw*0602 and Cw*0702 (06mut and 07mut, respectively) are aligned to 3′UTR sequences of native Cw*0602 and Cw*0702. Identical nucleotides are shown as dots, altered nucleotides are underlined, and deletions are indicated by hyphens (−) for optimal alignment. b. Fold change in luciferase activity of the modified 3′UTR (06mut and 07mut). c. Fold change in luciferase activity of reporters containing wild type Cw*0602 or Cw*0702 3′UTR sequences upon introduction of miR-148a mimic and inhibitor. d. Fold change in luciferase activity of reporters containing 06mut and 07mut 3′UTR sequences upon introduction of miR-148a mimic and inhibitor. Presence (+) or absence (−) of each variable, including a negative control (NC) miRNA, a mimic of miR-148a, or an inhibitor of miR-148a is shown. The data represent six replicates in each experimental group, the Mean ±SE are depicted as horizontal and vertical bars for each group, respectively, and one of three comparable experiments performed is shown. Non-parametric Wilcoxon-Mann-Whitney tests were used for statistical comparisons and two tailed p values are indicated.
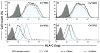
Figure 3. miR-148a affects cell surface expression of HLA-C
Histograms of HLA-C cell surface expression on HLA-C homozygous BLCL using flow cytometry are illustrated. In each plot, a NC miRNA that does not bind to the 3′UTR of HLA-C was included. a–b. HLA-Cw*0602 homozygous cells (BLCL-Cw*0602Hom) transfected with either a mimic or an inhibitor of miR-148a (a) or miR-148b (b). c–d. HLA-Cw*0702 homozygous cells (BLCL-Cw*0702Hom) transfected with either a mimic or an inhibitor of miR-148a (c) or miR-148b (d).
Similar articles
- Genetic interplay between HLA-C and MIR148A in HIV control and Crohn disease.
Kulkarni S, Qi Y, O'hUigin C, Pereyra F, Ramsuran V, McLaren P, Fellay J, Nelson G, Chen H, Liao W, Bass S, Apps R, Gao X, Yuki Y, Lied A, Ganesan A, Hunt PW, Deeks SG, Wolinsky S, Walker BD, Carrington M. Kulkarni S, et al. Proc Natl Acad Sci U S A. 2013 Dec 17;110(51):20705-10. doi: 10.1073/pnas.1312237110. Epub 2013 Nov 18. Proc Natl Acad Sci U S A. 2013. PMID: 24248364 Free PMC article. Clinical Trial. - High frequency of HIV mutations associated with HLA-C suggests enhanced HLA-C-restricted CTL selective pressure associated with an AIDS-protective polymorphism.
Blais ME, Zhang Y, Rostron T, Griffin H, Taylor S, Xu K, Yan H, Wu H, James I, John M, Dong T, Rowland-Jones SL. Blais ME, et al. J Immunol. 2012 May 1;188(9):4663-70. doi: 10.4049/jimmunol.1103472. Epub 2012 Apr 2. J Immunol. 2012. PMID: 22474021 Free PMC article. Clinical Trial. - The molecular origin and consequences of escape from miRNA regulation by HLA-C alleles.
O'huigin C, Kulkarni S, Xu Y, Deng Z, Kidd J, Kidd K, Gao X, Carrington M. O'huigin C, et al. Am J Hum Genet. 2011 Sep 9;89(3):424-31. doi: 10.1016/j.ajhg.2011.07.024. Am J Hum Genet. 2011. PMID: 21907013 Free PMC article. - The emerging role of HLA-C in HIV-1 infection.
Kulpa DA, Collins KL. Kulpa DA, et al. Immunology. 2011 Oct;134(2):116-22. doi: 10.1111/j.1365-2567.2011.03474.x. Immunology. 2011. PMID: 21896007 Free PMC article. Review. - HLA-C and HIV-1: friends or foes?
Zipeto D, Beretta A. Zipeto D, et al. Retrovirology. 2012 May 9;9:39. doi: 10.1186/1742-4690-9-39. Retrovirology. 2012. PMID: 22571741 Free PMC article. Review.
Cited by
- Epigenetic regulation of differential HLA-A allelic expression levels.
Ramsuran V, Kulkarni S, O'huigin C, Yuki Y, Augusto DG, Gao X, Carrington M. Ramsuran V, et al. Hum Mol Genet. 2015 Aug 1;24(15):4268-75. doi: 10.1093/hmg/ddv158. Epub 2015 May 1. Hum Mol Genet. 2015. PMID: 25935001 Free PMC article. - Nucleotide alterations in the HLA-C class I gene can cause aberrant splicing and marked changes in RNA levels in a polymorphic context-dependent manner.
Mizutani A, Suzuki S, Shigenari A, Sato T, Tanaka M, Kulski JK, Shiina T. Mizutani A, et al. Front Immunol. 2024 Jan 24;14:1332636. doi: 10.3389/fimmu.2023.1332636. eCollection 2023. Front Immunol. 2024. PMID: 38327766 Free PMC article. - Unravelling the mechanisms of durable control of HIV-1.
Walker BD, Yu XG. Walker BD, et al. Nat Rev Immunol. 2013 Jul;13(7):487-98. doi: 10.1038/nri3478. Nat Rev Immunol. 2013. PMID: 23797064 Review. - Natural killer cells in healthy and diseased subjects.
Biassoni R, Coligan JE, Moretta L. Biassoni R, et al. J Biomed Biotechnol. 2011;2011:795251. doi: 10.1155/2011/795251. Epub 2011 Aug 29. J Biomed Biotechnol. 2011. PMID: 21904444 Free PMC article. No abstract available. - Genetic association of human leukocyte antigens with chronicity or resolution of hepatitis B infection in thai population.
Posuwan N, Payungporn S, Tangkijvanich P, Ogawa S, Murakami S, Iijima S, Matsuura K, Shinkai N, Watanabe T, Poovorawan Y, Tanaka Y. Posuwan N, et al. PLoS One. 2014 Jan 23;9(1):e86007. doi: 10.1371/journal.pone.0086007. eCollection 2014. PLoS One. 2014. PMID: 24465836 Free PMC article.
References
- Snary D, Barnstable CJ, Bodmer WF, Crumpton MJ. Molecular structure of human histocompatibility antigens: the HLA-C series. Eur J Immunol. 1977;7:580–585. - PubMed
- Bashirova AA, Martin MP, McVicar DW, Carrington M. The killer immunoglobulin-like receptor gene cluster: tuning the genome for defense. Annu Rev Genomics Hum Genet. 2006;7:277–300. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- U01 AI035042/AI/NIAID NIH HHS/United States
- 5-M01-RR-00722/RR/NCRR NIH HHS/United States
- U01-AI-35042/AI/NIAID NIH HHS/United States
- U01-AI-37613/AI/NIAID NIH HHS/United States
- R01-DA04334/DA/NIDA NIH HHS/United States
- R01 DA004334/DA/NIDA NIH HHS/United States
- UL1 RR024131/RR/NCRR NIH HHS/United States
- M01 RR000722/RR/NCRR NIH HHS/United States
- U01 AI035041/AI/NIAID NIH HHS/United States
- R01-DA12568/DA/NIDA NIH HHS/United States
- P30 AI027763/AI/NIAID NIH HHS/United States
- U01 AI035043/AI/NIAID NIH HHS/United States
- U01 AI035040/AI/NIAID NIH HHS/United States
- HHSN261200800001C/RC/CCR NIH HHS/United States
- HHSN261200800001E/CA/NCI NIH HHS/United States
- U01 AI037984/AI/NIAID NIH HHS/United States
- U01-AI-35043/AI/NIAID NIH HHS/United States
- U01 AI037613/AI/NIAID NIH HHS/United States
- U01-AI-35041/AI/NIAID NIH HHS/United States
- U01-AI-35040/AI/NIAID NIH HHS/United States
- ImNIH/Intramural NIH HHS/United States
- U01-AI-37984/AI/NIAID NIH HHS/United States
- N02 CP055504/CP/NCI NIH HHS/United States
- R01 DA012568/DA/NIDA NIH HHS/United States
- U01-AI-35039/AI/NIAID NIH HHS/United States
- U01 AI035039/AI/NIAID NIH HHS/United States
- P30 AI060354/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials