MDAnalysis: a toolkit for the analysis of molecular dynamics simulations - PubMed (original) (raw)
. 2011 Jul 30;32(10):2319-27.
doi: 10.1002/jcc.21787. Epub 2011 Apr 15.
Affiliations
- PMID: 21500218
- PMCID: PMC3144279
- DOI: 10.1002/jcc.21787
MDAnalysis: a toolkit for the analysis of molecular dynamics simulations
Naveen Michaud-Agrawal et al. J Comput Chem. 2011.
Abstract
MDAnalysis is an object-oriented library for structural and temporal analysis of molecular dynamics (MD) simulation trajectories and individual protein structures. It is written in the Python language with some performance-critical code in C. It uses the powerful NumPy package to expose trajectory data as fast and efficient NumPy arrays. It has been tested on systems of millions of particles. Many common file formats of simulation packages including CHARMM, Gromacs, Amber, and NAMD and the Protein Data Bank format can be read and written. Atoms can be selected with a syntax similar to CHARMM's powerful selection commands. MDAnalysis enables both novice and experienced programmers to rapidly write their own analytical tools and access data stored in trajectories in an easily accessible manner that facilitates interactive explorative analysis. MDAnalysis has been tested on and works for most Unix-based platforms such as Linux and Mac OS X. It is freely available under the GNU General Public License from http://mdanalysis.googlecode.com.
Keywords: Python programming language; analysis; membrane systems; molecular dynamics simulations; object-oriented design; proteins; software.
Copyright © 2011 Wiley Periodicals, Inc.
Figures
Figure 1
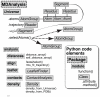
Organization of the MDAnalysis Python library. The Universe class contains both topological and structural information and maintains a list of all atoms – an AtomGroup instance named atoms – in the system. The selectAtoms() methods provides an interface to the selection mechanism and returns an AtomGroup instance. A molecular dynamics trajectory is accessed through the trajectory attribute, which is an instance of a Reader class. MDAnalysis also contains an analysis sub-module, which collects a number of pre-defined classes and tools.
Figure 2
Layout of important MDAnalysis classes. The Universe class contains an AtomGroup of all Atom instances that can be accessed through the attribute Universe.atoms. It also features a number of auto-generated _AtomGroup_s, one for each segment identifier of the topology. Users can obtain information from the individual Atom instances and computed data from a whole AtomGroup by querying the instance attributes and methods. The Timestep class represents the current state of the system at a particular time frame of the trajectory. Trajectory Reader and Writer classes provide an abstract interface to trajectory I/O. They implement special methods that allow “Pythonic” access to the objects such as treating a Reader as an iterator over trajectory frames (via the __ iter__ () special method) or selecting individual frames by the indexing operation (implemented through a custom __ getitem__ () method).
Figure 3
Calculation of geometric parameters for a KALP19 peptide in a DMPC bilayer. (A) The alpha-helical KALP19 peptide (black) was simulated in a DMPC bilayer with 90 lipids with the CHARMM27 force field. (B) Backbone dihedral angles φ and ψ averaged over a 75 ns MD trajectory. Error bars indicate one standard deviation from the mean. The values for an ideal α-helix are shown as dashed lines (φ = −57° and ψ = −47°) for comparison. (C) Average deuterium order parameter profiles, _S_CD, of DMPC lipid tails. _S_CD converges slowly and only the average over the last 25 ns is shown.
Figure 4
Defining membrane leaflets (MDAnalysis.analysis.leaflets.LeafletFinder). A-C: LeafletFinder algorithm (A) A graph is constructed that connects particles within a fixed cutoff (circles) such as lipid head group particles. (B) An algorithm implemented in the NetworkX package detects all disconnected subgraphs. (C) Typically, only two graphs are found that describe the two topologically disjoint leaflets. (D) View of a coarse grained bilayer with strong deformations. The bilayer contains ~24,000 lipids and the total simulation system size was ~1.5 million particles (J. Goose, personal communication). The phosphate particles in the head groups are shown and are colored black or white according to the LeafletFinder algorithm. (E) Close-up view of a deformation that is larger than the bilayer thickness itself.
Figure 5
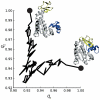
Native contacts analysis for the closed to open transition of adenylate kinase. _q_1 is the fraction of native contacts relative to the closed state (PDB: 1AKE) and _q_2 is relative to an open state structure (PDB: 4AKE).
Similar articles
- LOOS: an extensible platform for the structural analysis of simulations.
Romo TD, Grossfield A. Romo TD, et al. Annu Int Conf IEEE Eng Med Biol Soc. 2009;2009:2332-5. doi: 10.1109/IEMBS.2009.5335065. Annu Int Conf IEEE Eng Med Biol Soc. 2009. PMID: 19965179 - Morphoscanner2.0: A new python module for analysis of molecular dynamics simulations.
Fontana F, Carlino C, Malik A, Gelain F. Fontana F, et al. PLoS One. 2023 Apr 27;18(4):e0284307. doi: 10.1371/journal.pone.0284307. eCollection 2023. PLoS One. 2023. PMID: 37104393 Free PMC article. - Pytim: A python package for the interfacial analysis of molecular simulations.
Sega M, Hantal G, Fábián B, Jedlovszky P. Sega M, et al. J Comput Chem. 2018 Sep 30;39(25):2118-2125. doi: 10.1002/jcc.25384. Epub 2018 Oct 10. J Comput Chem. 2018. PMID: 30306571 Free PMC article. - Analysis Libraries for Molecular Trajectories: A Cross-Language Synopsis.
Giorgino T. Giorgino T. Methods Mol Biol. 2019;2022:503-527. doi: 10.1007/978-1-4939-9608-7_20. Methods Mol Biol. 2019. PMID: 31396916 Review. - Array programming with NumPy.
Harris CR, Millman KJ, van der Walt SJ, Gommers R, Virtanen P, Cournapeau D, Wieser E, Taylor J, Berg S, Smith NJ, Kern R, Picus M, Hoyer S, van Kerkwijk MH, Brett M, Haldane A, Del Río JF, Wiebe M, Peterson P, Gérard-Marchant P, Sheppard K, Reddy T, Weckesser W, Abbasi H, Gohlke C, Oliphant TE. Harris CR, et al. Nature. 2020 Sep;585(7825):357-362. doi: 10.1038/s41586-020-2649-2. Epub 2020 Sep 16. Nature. 2020. PMID: 32939066 Free PMC article. Review.
Cited by
- Dissected antiporter modules establish minimal proton-conduction elements of the respiratory complex I.
Beghiah A, Saura P, Badolato S, Kim H, Zipf J, Auman D, Gamiz-Hernandez AP, Berg J, Kemp G, Kaila VRI. Beghiah A, et al. Nat Commun. 2024 Oct 22;15(1):9098. doi: 10.1038/s41467-024-53194-5. Nat Commun. 2024. PMID: 39438463 Free PMC article. - Membrane association of the PTEN tumor suppressor: electrostatic interaction with phosphatidylserine-containing bilayers and regulatory role of the C-terminal tail.
Shenoy SS, Nanda H, Lösche M. Shenoy SS, et al. J Struct Biol. 2012 Dec;180(3):394-408. doi: 10.1016/j.jsb.2012.10.003. Epub 2012 Oct 13. J Struct Biol. 2012. PMID: 23073177 Free PMC article. - Detecting the First Hydration Shell Structure around Biomolecules at Interfaces.
Konstantinovsky D, Perets EA, Santiago T, Velarde L, Hammes-Schiffer S, Yan ECY. Konstantinovsky D, et al. ACS Cent Sci. 2022 Oct 26;8(10):1404-1414. doi: 10.1021/acscentsci.2c00702. Epub 2022 Sep 6. ACS Cent Sci. 2022. PMID: 36313165 Free PMC article. - Lipid-mediated activation of plasma membrane-localized deubiquitylating enzymes modulate endosomal trafficking.
Vogel K, Bläske T, Nagel MK, Globisch C, Maguire S, Mattes L, Gude C, Kovermann M, Hauser K, Peter C, Isono E. Vogel K, et al. Nat Commun. 2022 Nov 12;13(1):6897. doi: 10.1038/s41467-022-34637-3. Nat Commun. 2022. PMID: 36371501 Free PMC article. - PyMM: An Open-Source Python Program for QM/MM Simulations Based on the Perturbed Matrix Method.
Chen CG, Nardi AN, Amadei A, D'Abramo M. Chen CG, et al. J Chem Theory Comput. 2023 Jan 10;19(1):33-41. doi: 10.1021/acs.jctc.2c00767. Epub 2022 Nov 15. J Chem Theory Comput. 2023. PMID: 36378163 Free PMC article.
References
- Hess B, Kutzner C, van der Spoel D, Lindahl E. J Chem Theo Comp. 2008;4(3):435–447. - PubMed
- Seeber M, Cecchini M, Rao F, Settanni G, Caflisch A. Bioinformatics. 2007;23(19):2625–2627. - PubMed
- Verstraelen T, Van Houteghem M, Van Speybroeck V, Waroquier M. J Chem Inform Model. 2008;48(12):2414–2424. - PubMed
- Mezei M. J Comput Chem. 2010;31(14):2658–2668. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources