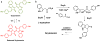The biosynthesis of cyanobacterial sunscreen scytonemin in intertidal microbial mat communities - PubMed (original) (raw)
The biosynthesis of cyanobacterial sunscreen scytonemin in intertidal microbial mat communities
Emily P Balskus et al. FEMS Microbiol Ecol. 2011 Aug.
Abstract
We have examined the biosynthesis and accumulation of cyanobacterial sunscreening pigment scytonemin within intertidal microbial mat communities using a combination of chemical, molecular, and phylogenetic approaches. Both laminated (layered) and nonlaminated mats contained scytonemin, with morphologically distinct mats having different cyanobacterial community compositions. Within laminated microbial mats, regions with and without scytonemin had different dominant oxygenic phototrophs, with scytonemin-producing areas consisting primarily of Lyngbya aestuarii and scytonemin-deficient areas dominated by a eukaryotic alga. The nonlaminated mat was populated by a diverse group of cyanobacteria and did not contain algae. The amplification and phylogenetic assignment of scytonemin biosynthetic gene scyC from laminated mat samples confirmed that the dominant cyanobacterium in these areas, L. aestuarii, is likely responsible for sunscreen production. This study is the first to utilize an understanding of the molecular basis of scytonemin assembly to explore its synthesis and function within natural microbial communities.
© 2011 Federation of European Microbiological Societies. Published by Blackwell Publishing Ltd. All rights reserved.
Figures
Figure 1
A) The structures of scytonemin and reduced scytonemin. B) Proposed biosynthetic pathway for scytonemin production with biochemically characterized enzymatic transformations.
Figure 2
Photographs of Little Sippewissett microbial mats. A) Mat 1 B) Detail of Mat 1 surface with dark green, leathery scytonemin-producing areas and lighter green, felt-like areas without scytonemin. C) Mat 5 D) Mat 6 E) Isolated scytonemin from mat samples.
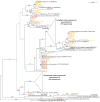
Figure 3
Maximum likelihood phylogeny of 16S rRNA gene sequences from microbial mats. The tree was compiled by maximum likelihood using PHYML. Bootstrap confidence values >50 are indicated on the nodes. Strains are indicated after species names, followed by the accession number in parenthesis. Sequences are colored according to library (red = Mat 1A [scytonemin producing area], green = Mat 1B [scytonemin deficient area], blue = Mat 5, orange = Mat 6).
Figure 4
Maximum likelihood phylogeny of A) Lyngbya and B) chloroplast 16S rRNA gene sequences from microbial mats. The tree was compiled by maximum likelihood using PHYML. Bootstrap confidence values >50 are indicated on the nodes. Strains are indicated after species names, followed by the accession number in parenthesis.
Figure 4
Maximum likelihood phylogeny of A) Lyngbya and B) chloroplast 16S rRNA gene sequences from microbial mats. The tree was compiled by maximum likelihood using PHYML. Bootstrap confidence values >50 are indicated on the nodes. Strains are indicated after species names, followed by the accession number in parenthesis.
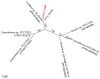
Figure 5
Richness and diversity analyses of microbial mat 16S rRNA gene clone libraries. The operational taxonomic unit definition used in all analyses corresponds to 98% DNA sequence identity (OTU0.02). A) UPGMA dendrogram of the similarity between clone libraries (including cyanobacterial and chloroplastic sequences). B) Venn diagram representing the number of OTU0.02 shared by the clone libraries (including cyanobacterial and chloroplastic sequences). The total number of OTUs in each library is shown in brackets. C) Rarefaction curves for the cyanobacterial sequences of the three largest clone libraries. Error bars represent a 95% confidence interval.
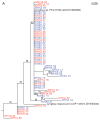
Figure 6
Maximum likelihood phylogenetic tree of translated ScyC amino acid sequences from sequenced cyanobacteria and Mats 1 and 5. The tree was compiled by maximum likelihood using PHYML. Bootstrap confidence values >50 are indicated on the nodes. The number in parentheses found after each taxon name is the accession number for the respective protein sequence.
Similar articles
- Lipid biomarkers, pigments and cyanobacterial diversity of microbial mats across intertidal flats of the arid coast of the Arabian Gulf (Abu Dhabi, UAE).
Abed RM, Kohls K, Schoon R, Scherf AK, Schacht M, Palinska KA, Al-Hassani H, Hamza W, Rullkötter J, Golubic S. Abed RM, et al. FEMS Microbiol Ecol. 2008 Sep;65(3):449-62. doi: 10.1111/j.1574-6941.2008.00537.x. Epub 2008 Jul 11. FEMS Microbiol Ecol. 2008. PMID: 18637042 - A comparative genomics approach to understanding the biosynthesis of the sunscreen scytonemin in cyanobacteria.
Soule T, Palmer K, Gao Q, Potrafka RM, Stout V, Garcia-Pichel F. Soule T, et al. BMC Genomics. 2009 Jul 24;10:336. doi: 10.1186/1471-2164-10-336. BMC Genomics. 2009. PMID: 19630972 Free PMC article. - Biotechnological Production of the Sunscreen Pigment Scytonemin in Cyanobacteria: Progress and Strategy.
Gao X, Jing X, Liu X, Lindblad P. Gao X, et al. Mar Drugs. 2021 Feb 27;19(3):129. doi: 10.3390/md19030129. Mar Drugs. 2021. PMID: 33673485 Free PMC article. Review. - Bioinformatic, phylogenetic and chemical analysis of the UV-absorbing compounds scytonemin and mycosporine-like amino acids from the microbial mat communities of Shark Bay, Australia.
D'Agostino PM, Woodhouse JN, Liew HT, Sehnal L, Pickford R, Wong HL, Burns BP, Neilan BA. D'Agostino PM, et al. Environ Microbiol. 2019 Feb;21(2):702-715. doi: 10.1111/1462-2920.14517. Epub 2019 Jan 25. Environ Microbiol. 2019. PMID: 30589201 - Cyanobacterial Sunscreen Scytonemin: Role in Photoprotection and Biomedical Research.
Rastogi RP, Sonani RR, Madamwar D. Rastogi RP, et al. Appl Biochem Biotechnol. 2015 Jul;176(6):1551-63. doi: 10.1007/s12010-015-1676-1. Epub 2015 May 27. Appl Biochem Biotechnol. 2015. PMID: 26013282 Review.
Cited by
- Molecular musings in microbial ecology and evolution.
Case RJ, Boucher Y. Case RJ, et al. Biol Direct. 2011 Nov 10;6:58. doi: 10.1186/1745-6150-6-58. Biol Direct. 2011. PMID: 22074255 Free PMC article. Review. - Gypsum endolithic phototrophs under moderate climate (Southern Sicily): their diversity and pigment composition.
Němečková K, Mareš J, Procházková L, Culka A, Košek F, Wierzchos J, Nedbalová L, Dudák J, Tymlová V, Žemlička J, Kust A, Zima J, Nováková E, Jehlička J. Němečková K, et al. Front Microbiol. 2023 Jul 6;14:1175066. doi: 10.3389/fmicb.2023.1175066. eCollection 2023. Front Microbiol. 2023. PMID: 37485515 Free PMC article. - Influence of Low Salt Concentration on Growth Behavior and General Biomass Composition in Lyngbya purpurem (Cyanobacteria).
López-Pacheco IY, Fuentes-Tristan S, Rodas-Zuluaga LI, Castillo-Zacarías C, Pedro-Carrillo I, Martínez-Prado MA, Iqbal HMN, Parra-Saldívar R. López-Pacheco IY, et al. Mar Drugs. 2020 Dec 4;18(12):621. doi: 10.3390/md18120621. Mar Drugs. 2020. PMID: 33291783 Free PMC article. - Benthic cyanobacterial mats in the high arctic: multi-layer structure and fluorescence responses to osmotic stress.
Lionard M, Péquin B, Lovejoy C, Vincent WF. Lionard M, et al. Front Microbiol. 2012 Apr 26;3:140. doi: 10.3389/fmicb.2012.00140. eCollection 2012. Front Microbiol. 2012. PMID: 22557996 Free PMC article. - Inferred ancestry of scytonemin biosynthesis proteins in cyanobacteria indicates a response to Paleoproterozoic oxygenation.
Tamre E, Fournier GP. Tamre E, et al. Geobiology. 2022 Nov;20(6):764-775. doi: 10.1111/gbi.12514. Epub 2022 Jul 18. Geobiology. 2022. PMID: 35851984 Free PMC article.
References
- Branowitz S, Castenholz R. Long-term effects of UV and visible irradiance on natural populations of a scytonemin-containing cyanobacteriam (Calothrix sp.) FEMS Microbiol Ecol. 1997;24:343–352.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases