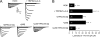Trafficking and gating of hyperpolarization-activated cyclic nucleotide-gated channels are regulated by interaction with tetratricopeptide repeat-containing Rab8b-interacting protein (TRIP8b) and cyclic AMP at distinct sites - PubMed (original) (raw)
Trafficking and gating of hyperpolarization-activated cyclic nucleotide-gated channels are regulated by interaction with tetratricopeptide repeat-containing Rab8b-interacting protein (TRIP8b) and cyclic AMP at distinct sites
Ye Han et al. J Biol Chem. 2011.
Abstract
Ion channel trafficking and gating are often influenced by interactions with auxiliary subunits. Tetratricopeptide repeat-containing Rab8b-interacting protein (TRIP8b) is an auxiliary subunit for neuronal hyperpolarization-activated cyclic nucleotide-gated (HCN) channels. TRIP8b interacts directly with two distinct sites of HCN channel pore-forming subunits to control channel trafficking and gating. Here we use mutagenesis combined with electrophysiological studies to define and distinguish the functional importance of the HCN/TRIP8b interaction sites. Interaction with the last three amino acids of the HCN1 C terminus governed the effect of TRIP8b on channel trafficking, whereas TRIP8b interaction with the HCN1 cyclic nucleotide binding domain (CNBD) affected trafficking and gating. Biochemical studies revealed that direct interaction between TRIP8b and the HCN1 CNBD was disrupted by cAMP and that TRIP8b binding to the CNBD required an arginine residue also necessary for cAMP binding. In accord, increasing cAMP levels in cells antagonized the up-regulation of HCN1 channels mediated by a TRIP8b construct binding the CNBD exclusively. These data illustrate the distinct roles of the two TRIP8b-HCN interaction domains and suggest that TRIP8b and cAMP may directly compete for binding the HCN CNBD to control HCN channel gating, kinetics, and trafficking.
Figures
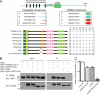
FIGURE 1.
TRIP8b interacts with HCN channels at two distinct sites. A, schematic representations of HCN subunit and TRIP8b constructs tested in directed yeast two-hybrid interactions are shown. The + indicates auxotrophic growth resulting from interaction of bait and prey plasmids, whereas 0 signifies no interaction. B, Western blots of extracts from HEK293T cells transfected with HCN1 and the indicated TRIP8b constructs were immunoprecipitated with α-TRIP8b and probed with α-HCN1 (top panels) or α-TRIP8b (bottom panels). Input is 6% that of the total lysate, whereas immunoprecipitate (IP) is 15%. Molecular mass markers are shown in kDa. C, shown are summary data for interaction of HCN1 and TRIP8b. ** and ***, p < 0.01 and 0.001, respectively, versus TRIP8b(1a2–4), 1-way ANOVA with Tukey's post hoc test, n = 3).
FIGURE 2.
TRIP8b interaction with the HCN1 CNBD and the C-terminal tripeptide contribute to TRIP8b-mediated increases in HCN1 current density. A, example Ih traces recorded from transfected HEK293 cells in response to a series of hyperpolarizing steps (from −40 to −130 mV in decrements of −10 mV) are shown. Cells were transfected with HCN1 alone (left trace) or co-transfected with both HCN1 and TRIP8b(1a-2-4) mutants as indicated. B, shown is a quantitative summary of Ih density in HEK293 cells co-transfected with HCN1 and TRIP8b(1a-2-4) mutants. ** and ***, p < 0.01 and 0.001, respectively, compared with the current density of HCN1 when expressed alone. ++ and +++, p < 0.01 and 0.001, respectively, compared with currents obtained upon transfection of HCN1 with wild type TRIP8b(1a-2-4). No. of cells is indicated per experimental group on the bar histogram.
FIGURE 3.
TRIP8b interaction with either the HCN1 CNBD or the C-terminal tripeptide is required for TRIP8b-mediated increases in HCN1 surface expression. A, shown are representative flow cytometric histograms obtained for cells labeled with surface-HA when a construct encoding HCN1-HA was co-expressed with eGFP alone or with eGFP and the indicated TRIP8b(1a-2-4) constructs. The x axis indicates the fluorescence intensity in arbitrary units (note the log scale), and the y axis indicates the percentage of the maximum cell count. Representative distributions obtained from eGFP-transfected cells (control) were overlaid with each experimental condition as indicated. B, summary data for flow cytometric studies of surface expression of HCN1-HA are shown. Compared with HCN1 alone, TRIP8b(1a-2-4), TRIP8b(1a-2-4)[TPR3-N13A], and TRIP8b(1a-2-4)[Δ58] significantly increased the HCN1 surface expression, whereas TRIP8b(1a-2-4)[Δ58+TPR3-N13A] had no effect (***, p < 0.001; *, p < 0.05, n = 6). C, representative Western blots of extracts from HEK293T cells transfected with HCN1 and the indicated TRIP8b constructs. Surface HA epitope was bound in non-permeabilized cells before solubilization and immunoprecipitation with α-HA. D, summary data for surface HCN1-HA immunoprecipitation are shown. Compared with HCN1 alone, wild type TRIP8b(1a-2-4), TRIP8b(1a-2-4)[TPR3-N13A], and TRIP8b(1a-2-4)[Δ58] significantly increased the amount of HCN1 surface expression, whereas TRIP8b(1a-2-4)[Δ58+TPR3-N13A] had no effect (***, p < 0.001, 1-way ANOVA with Tukey's post hoc-test, n = 3). Input was 6% of total lysate, and immunoprecipitation was 15%. Molecular mass markers are shown in kDa.
FIGURE 4.
Slowing of HCN1 activation kinetics by TRIP8b is mediated exclusively by interaction with the CNBD interaction domain. A–D, Ih activation kinetics in HEK293 cells expressing HCN1 and various mutants of TRIP8b(1a-2-4) are shown. Gray data points represent the activation kinetics of HCN1 when transfected without TRIP8b (the same in all four panels), and black data points represent the activation kinetics when HCN1 was co-transfected with various TRIP8b(1a-2-4) mutants, as indicated. Insets are example traces comparing Ih (evoked by a hyperpolarizing step to −110 mV) in the presence (black) and absence (gray) of the indicated TRIP8b mutant. Only the first 500 ms of the response are shown. E, comparison of all traces is shown in the insets to panel A-D. The currents were normalized to the amplitude at the end of the pulse (3 s) and are presented at logarithmic scale for demonstration purposes. Statistical comparison of time constants at specific voltages to their counterparts in control cells was performed using ANOVA followed by Dunnett's post hoc test; n = 6–15 cells per data point. * = p < 0.05; ** = p < 0.01.
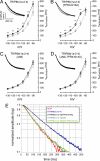
FIGURE 5.
The hyperpolarizing shift in the voltage activation of HCN1 channels by TRIP8b is mediated solely by interaction of HCN1 with the CNBD interaction domain. A, activation curves are based on Ih recorded in HEK293 cells expressing HCN1 together with different TRIP8b mutants as indicated. Deletion of the CNBD-interacting domain (Δ58) resulted in a voltage activation curve not different from the one obtained when HCN1 is transfected alone, whereas interference with the C′-terminal -tripeptide interacting domain (TPR3-N13A) resulted in a hyperpolarizing shift of the activation curve similar to that obtained with wild type TRIP8b(1a-2-4). For illustration purposes, the curves presented in this figure are based on grouped and normalized data, whereas statistical analysis of _V_50 and the slope factor was performed on the parameters obtained from the individual Boltzmann fits. B, shown is quantification of the effects of mutations on the shift in half-maximal activation _V_50. See Table 3 for values and statistical significance.
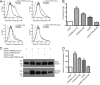
FIGURE 6.
cAMP disrupts TRIP8b-CNBD interactions and their functional effects. A and B, Western blots from GST pulldown experiments are shown. Input is 15% of lysate precipitated. A, extracts from HEK293T cells transfected with HCN1 (left panels) or HCN1Δ3 (right panels) were incubated with purified recombinant TRIP8b(1a)-GST alone or with the indicated concentration of cAMP, then precipitated with immobilized glutathione and probed for HCN1 (top panels) or TRIP8b (bottom panels). B, shown are summary data for cAMP disrupting TRIP8b-CNBD interaction. Values reflect density of bands corresponding to HCN1 constructs precipitated by TRIP8b(1a)-GST, normalized to conditions lacking cAMP. C, extracts from HEK293T cells transfected with HCN1, HCN1[Δ3], HCN1[R538E], or HCN1[Δ3+R538E] were incubated with purified recombinant GST alone, TRIP8b(1a)-GST, or TRIP8b(1c)-GST, then precipitated with immobilized glutathione and probed for HCN1. D, shown are summary data for flow cytometric studies of surface expression of HCN1-HA obtained for cells labeled with surface-HA. Cells were transfected with a construct encoding HCN1-HA and eGFP alone or with eGFP and the indicated TRIP8b(1a-2-4) constructs. Before flow cytometry, cells were incubated with or without forskolin as indicated.
Similar articles
- The structure and function of TRIP8b, an auxiliary subunit of hyperpolarization-activated cyclic-nucleotide gated channels.
Han Y, Lyman KA, Foote KM, Chetkovich DM. Han Y, et al. Channels (Austin). 2020 Dec;14(1):110-122. doi: 10.1080/19336950.2020.1740501. Channels (Austin). 2020. PMID: 32189562 Free PMC article. Review. - TRIP8b regulates HCN1 channel trafficking and gating through two distinct C-terminal interaction sites.
Santoro B, Hu L, Liu H, Saponaro A, Pian P, Piskorowski RA, Moroni A, Siegelbaum SA. Santoro B, et al. J Neurosci. 2011 Mar 16;31(11):4074-86. doi: 10.1523/JNEUROSCI.5707-10.2011. J Neurosci. 2011. PMID: 21411649 Free PMC article. - Deletion of the hyperpolarization-activated cyclic nucleotide-gated channel auxiliary subunit TRIP8b impairs hippocampal Ih localization and function and promotes antidepressant behavior in mice.
Lewis AS, Vaidya SP, Blaiss CA, Liu Z, Stoub TR, Brager DH, Chen X, Bender RA, Estep CM, Popov AB, Kang CE, Van Veldhoven PP, Bayliss DA, Nicholson DA, Powell CM, Johnston D, Chetkovich DM. Lewis AS, et al. J Neurosci. 2011 May 18;31(20):7424-40. doi: 10.1523/JNEUROSCI.0936-11.2011. J Neurosci. 2011. PMID: 21593326 Free PMC article. - Structural basis for the mutual antagonism of cAMP and TRIP8b in regulating HCN channel function.
Saponaro A, Pauleta SR, Cantini F, Matzapetakis M, Hammann C, Donadoni C, Hu L, Thiel G, Banci L, Santoro B, Moroni A. Saponaro A, et al. Proc Natl Acad Sci U S A. 2014 Oct 7;111(40):14577-82. doi: 10.1073/pnas.1410389111. Epub 2014 Sep 2. Proc Natl Acad Sci U S A. 2014. PMID: 25197093 Free PMC article. - Neurophysiology of HCN channels: from cellular functions to multiple regulations.
He C, Chen F, Li B, Hu Z. He C, et al. Prog Neurobiol. 2014 Jan;112:1-23. doi: 10.1016/j.pneurobio.2013.10.001. Epub 2013 Oct 29. Prog Neurobiol. 2014. PMID: 24184323 Review.
Cited by
- Regulation of axonal HCN1 trafficking in perforant path involves expression of specific TRIP8b isoforms.
Wilkars W, Liu Z, Lewis AS, Stoub TR, Ramos EM, Brandt N, Nicholson DA, Chetkovich DM, Bender RA. Wilkars W, et al. PLoS One. 2012;7(2):e32181. doi: 10.1371/journal.pone.0032181. Epub 2012 Feb 21. PLoS One. 2012. PMID: 22363812 Free PMC article. - Reduced Hyperpolarization-Activated Current Contributes to Enhanced Intrinsic Excitability in Cultured Hippocampal Neurons from PrP(-/-) Mice.
Fan J, Stemkowski PL, Gandini MA, Black SA, Zhang Z, Souza IA, Chen L, Zamponi GW. Fan J, et al. Front Cell Neurosci. 2016 Mar 24;10:74. doi: 10.3389/fncel.2016.00074. eCollection 2016. Front Cell Neurosci. 2016. PMID: 27047338 Free PMC article. - The structure and function of TRIP8b, an auxiliary subunit of hyperpolarization-activated cyclic-nucleotide gated channels.
Han Y, Lyman KA, Foote KM, Chetkovich DM. Han Y, et al. Channels (Austin). 2020 Dec;14(1):110-122. doi: 10.1080/19336950.2020.1740501. Channels (Austin). 2020. PMID: 32189562 Free PMC article. Review. - Dynamic measurements for funny channels.
Puljung MC. Puljung MC. Proc Natl Acad Sci U S A. 2014 Oct 7;111(40):14320-1. doi: 10.1073/pnas.1416137111. Epub 2014 Sep 23. Proc Natl Acad Sci U S A. 2014. PMID: 25249634 Free PMC article. No abstract available. - Reelin signaling specifies the molecular identity of the pyramidal neuron distal dendritic compartment.
Kupferman JV, Basu J, Russo MJ, Guevarra J, Cheung SK, Siegelbaum SA. Kupferman JV, et al. Cell. 2014 Sep 11;158(6):1335-1347. doi: 10.1016/j.cell.2014.07.035. Epub 2014 Sep 4. Cell. 2014. PMID: 25201528 Free PMC article.
References
- Robinson R. B., Siegelbaum S. A. (2003) Annu. Rev. Physiol. 65, 453–480 - PubMed
- Biel M., Wahl-Schott C., Michalakis S., Zong X. (2009) Physiol. Rev. 89, 847–885 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- F30 NS064757/NS/NINDS NIH HHS/United States
- NS05595/NS/NINDS NIH HHS/United States
- NS35439/NS/NINDS NIH HHS/United States
- R37 NS035439/NS/NINDS NIH HHS/United States
- R25 GM079300/GM/NIGMS NIH HHS/United States
- NS064757/NS/NINDS NIH HHS/United States
- R01 NS059934/NS/NINDS NIH HHS/United States
- NS059934/NS/NINDS NIH HHS/United States
- R01 NS035439/NS/NINDS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases