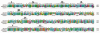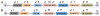Genome-wide comprehensive analysis of human helicases - PubMed (original) (raw)
Genome-wide comprehensive analysis of human helicases
Pavan Umate et al. Commun Integr Biol. 2011 Jan.
Abstract
Helicases are motor proteins that catalyze the unwinding of duplex nucleic acids in an ATP-dependent manner. They are involved in almost all the nucleic acid transactions. In the present study, we report a comprehensive analysis of helicase gene family in human and its comparison with homologs in model organisms. The human genome encodes for 95 non-redundant helicase proteins, of which 64 are RNA helicases and 31 are DNA helicases. 57 RNA helicases are validated based on annotations and occurrence of conserved helicase signature motifs. These include 14 DExH and 37 DExD subfamily members, six other members such as U5.snRNP, ATR-X, Suv3, FANCJ, and two of superkiller viralicidic activity 2-like helicases. 31 DNA helicases are also identified, which include RecQ, MCM and RuvB-like helicases. Finding a set of helicases in human and almost similar sequences in model organisms suggests that the "core" members of helicase gene family are highly conserved throughout evolution. The present study gives an overview of members of RNA and DNA helicases encoded by the human genome along with their conserved motifs, phylogeny and homologs in model organisms. The study on comparing these homologs will spread light on the organization and complexity of helicase gene family in model organisms. The comprehensive analysis of human helicases presented in this study will further provide an invaluable resource for elaborate biological research on these helicases.
Keywords: DEAD-box; DHX and DDX helicases; MCM proteins; RNA helicases; RecQ helicases; human helicases.
Figures
Figure 1
Protein alignment of 14 DHX family members of RNA helicases. (A) Entire protein sequence is aligned with ClustalX software. The eight conserved helicase signature motifs are identified on top of the alignment. The names of DHX helicases are identified on the left and sequence positions are given on right. Asterisks and dots drawn on top of sequence indicate identical residues and conservative amino acid changes, respectively. Gaps in the amino acid sequences are introduced to improve the alignment. Only a part of protein alignment with conserved helicase motifs is shown. (B) Schematic representations for DHX32 and DHX58 helicase motifs are shown. Note the presence of DExH motif in DDX members like DDX11, DDX12, DDX58 and DDX60. The typical motifs for DHX-type family members are represented on top.
Figure 2A and B
Protein alignment of 37 DDX family members of RNA helicases. (A) Alignment is made on entire protein sequences with ClustalX software. The nine conserved helicase motifs are highlighted on top of the alignment. The names of DDX helicases are identified on the left and sequence positions are given on right. Asterisks and dots drawn on top of sequence indicate identical residues and conservative amino acid changes, respectively. Gaps in the amino acid sequences are introduced to improve the alignment. Only a part of protein alignment with conserved helicase motifs is shown. (B) Diagrammatic representation of helicase motifs for typical DDX-type helicases and DDX1 are shown.
Figure 2A and B
Protein alignment of 37 DDX family members of RNA helicases. (A) Alignment is made on entire protein sequences with ClustalX software. The nine conserved helicase motifs are highlighted on top of the alignment. The names of DDX helicases are identified on the left and sequence positions are given on right. Asterisks and dots drawn on top of sequence indicate identical residues and conservative amino acid changes, respectively. Gaps in the amino acid sequences are introduced to improve the alignment. Only a part of protein alignment with conserved helicase motifs is shown. (B) Diagrammatic representation of helicase motifs for typical DDX-type helicases and DDX1 are shown.
Figure 3
Protein alignment of superkiller helicase group members, SKI2W and SKIV2L2. The consensus for eight conserved motifs, motif I (AHTSAGKT), motif Ia (TSPIKALSNQ), motif Ib (MTTE IL), motif II (DExH), motif III (SAT), motif IV (xxFTFSx), motif V (TET FAMGxNMPA) and motif VI (QMxGRA GR) are identified on top of the alignment made with ClustalX alignment program. The names of superkiller helicases are identified on the left and sequence positions are given on right. Asterisks and dots drawn on top of sequence indicate identical residues and conservative amino acid changes, respectively. Gaps in the amino acid sequences are introduced to improve the alignment. Only a part of protein alignment with conserved helicase motifs is shown.
Figure 4
Protein alignment of five RecQ helicases in human. The five RecQ members are identified on left and sequence position of amino acid residues are shown on right. The seven conserved RecQ motifs are labeled on top of the alignment. Asterisks and dots drawn on top of sequence indicate identical residues and conservative amino acid changes, respectively. Gaps in the amino acid sequences are introduced to improve the alignment. Only a part of protein alignment with conserved helicase motifs is shown.
Figure 5
Protein alignment of MCM proteins in human. Amino acid alignment of canonical MCM2-7, and the unusual MCM8 and MCM9 proteins. The figure is constructed with ClustalX program, names of MCM proteins are given on left, and sequence positions on right. Asterisks and dots drawn on top of sequence indicate identical residues and conservative amino acid changes, respectively. Gaps in the amino acid sequences are introduced to improve the alignment. Only a part of protein alignment with conserved helicase motifs is shown. Note the presence of a glycine (G) instead of alanine (A) or serine (S) in the Walker A motif of MCM8 and MCM9 proteins.
Figure 6
Schematic representation of seven conserved helicase motifs found in DNA repair helicases, XPB and XPD. The numbers given on top of the diagram indicate amino acid position of the first residue of a conserved motif for the entire protein sequence. (A) Amino acid consensus for motifs of XPB helicase. The sequences representing CGAGKS (motif I), VEQWK (motif Ia), LDEVH (motif II), LGLTATLxRED (motif III), PTSQGER (motif IV), FDLPEANV (motif V) and QRLGRVLR (motif VI) are shown. The first and last protein residues are methionine (M) and lysine (K) respectively are indicated. (B) Amino acid consensus for motifs of XPD helicase. The consensus sequence for these motifs is shown, SGTGKT (motif I), YCSRTV (motif Ia), FDEAH (motif II), TSGTLSPL (motif III), FTSYQYME (motif IV), SVARGKVS (motif V) and QCVGRAIR (motif VI). The first and last protein residues are methionine (M) and leucine (L) are shown.
Figure 7
Protein alignment of members of human ‘RuvB-like’ family of helicases. The Walker A (G(x)4GKT) and Walker B consensus (DExH) are indicated on top of the alignment. The sequences are aligned using ClustalX program. Names of RuvB-like proteins are given on left, and sequence positions on right. Asterisks and dots drawn on top of sequence indicate identical residues and conservative amino acid changes, respectively. Gaps in the amino acid sequences are introduced to improve the alignment. Only a part of protein alignment with conserved helicase motifs is shown.
Similar articles
- Identification and in silico analysis of cattle DExH/D box RNA helicases.
Suthar MK, Purva M, Maherchandani S, Kashyap SK. Suthar MK, et al. Springerplus. 2016 Jan 7;5:25. doi: 10.1186/s40064-015-1640-0. eCollection 2016. Springerplus. 2016. PMID: 26783509 Free PMC article. - The human DDX and DHX gene families of putative RNA helicases.
Abdelhaleem M, Maltais L, Wain H. Abdelhaleem M, et al. Genomics. 2003 Jun;81(6):618-22. doi: 10.1016/s0888-7543(03)00049-1. Genomics. 2003. PMID: 12782131 - Genome wide identification of Plasmodium falciparum helicases: a comparison with human host.
Tuteja R. Tuteja R. Cell Cycle. 2010 Jan 1;9(1):104-20. doi: 10.4161/cc.9.1.10241. Epub 2010 Jan 5. Cell Cycle. 2010. PMID: 20016272 - Prokaryotic and eukaryotic DNA helicases. Essential molecular motor proteins for cellular machinery.
Tuteja N, Tuteja R. Tuteja N, et al. Eur J Biochem. 2004 May;271(10):1835-48. doi: 10.1111/j.1432-1033.2004.04093.x. Eur J Biochem. 2004. PMID: 15128294 Free PMC article. Review. - General and Target-Specific DExD/H RNA Helicases in Eukaryotic Translation Initiation.
Shen L, Pelletier J. Shen L, et al. Int J Mol Sci. 2020 Jun 20;21(12):4402. doi: 10.3390/ijms21124402. Int J Mol Sci. 2020. PMID: 32575790 Free PMC article. Review.
Cited by
- Human RecQ helicases in DNA repair, recombination, and replication.
Croteau DL, Popuri V, Opresko PL, Bohr VA. Croteau DL, et al. Annu Rev Biochem. 2014;83:519-52. doi: 10.1146/annurev-biochem-060713-035428. Epub 2014 Mar 3. Annu Rev Biochem. 2014. PMID: 24606147 Free PMC article. Review. - DDX49 is an RNA helicase that affects translation by regulating mRNA export and the levels of pre-ribosomal RNA.
Awasthi S, Verma M, Mahesh A, K Khan MI, Govindaraju G, Rajavelu A, Chavali PL, Chavali S, Dhayalan A. Awasthi S, et al. Nucleic Acids Res. 2018 Jul 6;46(12):6304-6317. doi: 10.1093/nar/gky231. Nucleic Acids Res. 2018. PMID: 29618122 Free PMC article. - Recruitment, Duplex Unwinding and Protein-Mediated Inhibition of the Dead-Box RNA Helicase Dbp2 at Actively Transcribed Chromatin.
Ma WK, Paudel BP, Xing Z, Sabath IG, Rueda D, Tran EJ. Ma WK, et al. J Mol Biol. 2016 Mar 27;428(6):1091-1106. doi: 10.1016/j.jmb.2016.02.005. Epub 2016 Feb 11. J Mol Biol. 2016. PMID: 26876600 Free PMC article. - Putative SF2 helicases of the early-branching eukaryote Giardia lamblia are involved in antigenic variation and parasite differentiation into cysts.
Gargantini PR, Serradell MC, Torri A, Lujan HD. Gargantini PR, et al. BMC Microbiol. 2012 Nov 28;12:284. doi: 10.1186/1471-2180-12-284. BMC Microbiol. 2012. PMID: 23190735 Free PMC article. - Disease-Causing Mutations in SF3B1 Alter Splicing by Disrupting Interaction with SUGP1.
Zhang J, Ali AM, Lieu YK, Liu Z, Gao J, Rabadan R, Raza A, Mukherjee S, Manley JL. Zhang J, et al. Mol Cell. 2019 Oct 3;76(1):82-95.e7. doi: 10.1016/j.molcel.2019.07.017. Epub 2019 Aug 29. Mol Cell. 2019. PMID: 31474574 Free PMC article.
References
- Tuteja R. Genome wide identification of Plasmodium falciparum helicases: a comparison with human host. Cell Cycle. 2010;9:104–120. - PubMed
- Umate P, Tuteja R, Tuteja N. Genome-wide analysis of helicase gene family from rice and Arabidopsis: a comparison with yeast and human. Plant Mol Biol. 2010;73:449–465. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous