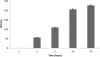Horizontal transfer of the OXA-24 carbapenemase gene via outer membrane vesicles: a new mechanism of dissemination of carbapenem resistance genes in Acinetobacter baumannii - PubMed (original) (raw)
Horizontal transfer of the OXA-24 carbapenemase gene via outer membrane vesicles: a new mechanism of dissemination of carbapenem resistance genes in Acinetobacter baumannii
Carlos Rumbo et al. Antimicrob Agents Chemother. 2011 Jul.
Abstract
The resistance of Acinetobacter baumannii strains to carbapenems is a worrying problem in hospital settings. The main mechanism of carbapenem resistance is the expression of β-lactamases (metalloenzymes or class D enzymes). The mechanisms of the dissemination of these genes among A. baumannii strains are not fully understood. In this study we used two carbapenem-resistant clinical strains of A. baumannii (AbH12O-A2 and AbH12O-CU3) expressing the plasmid-borne bla(OXA-24) gene (plasmids pMMA2 and pMMCU3, respectively) to demonstrate that A. baumannii releases outer membrane vesicles (OMVs) during in vitro growth. The use of hybridization studies enabled us to show that these OMVs harbored the bla(OXA-24) gene. The incubation of these OMVs with the carbapenem-susceptible A. baumannii ATCC 17978 host strain yielded full resistance to carbapenems. The presence of the original plasmids harboring the bla(OXA-24) gene was detected in strain ATCC 17978 after the transformation of OMVs. New OMVs harboring bla(OXA-24) were released by A. baumannii ATCC 17978 after it was transformed with the original OMV-mediated plasmids, indicating the universality of the process. We present the first experimental evidence that clinical isolates of A. baumannii may release OMVs as a mechanism of horizontal gene transfer whereby carbapenem resistance genes are delivered to surrounding A. baumannii bacterial isolates.
Figures
Fig. 1.
Electron microscopy micrograph of OMVs released by A. baumannii clinical strain AbH12O-A2. Vesicles were purified from broth cultures by ultracentrifugation and filtered through a 0.22-μm filter. The average diameter of the vesicles was 40 nm. The OMVs were free of contaminating bacteria.
Fig. 2.
Time-response experiment with supernatants from AbH12OA2 as a source of OMVs. Shown are numbers of colonies resistant to ampicillin (transformants) obtained at different times of incubation of A. baumannii ATCC 17978 with supernatant from AbH12OA2. At least three replicate experiments were performed, and each experiment had results similar to those shown. Error bars indicate the standard deviations for replicate samples.
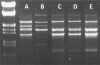
Fig. 3.
(A) Dose-response experiment. Shown is a dot blot analysis for detecting the presence of the _bla_OXA-24 gene in OMVs released from A. baumannii clinical isolate AbH12O-A2. Blot A, 10 μl of purified plasmid pMMA2 as a positive control; blot B, 10 μl of the OMV suspension (see concentration in Table 1); blot C, 50 μl of the OMV suspension; blot D, 100 μl of the OMV suspension; blot E, 200 μl of the OMV suspension; blot F, 100 μl of the OMV suspension from A. baumannii ATCC 17978 mock transformed as a negative control (equal amount of OMVs with respect to that in blot E). All samples were concentrated to a final volume of 10 μl before the experiment, and 5 μl of each sample was analyzed by dot blotting. (B) Dot blot analysis to detect the presence of the _bla_OXA-24 gene in OMVs released from A. baumannii clinical isolates AbH12O-A2 and AbH12O-CU3 (blots A and B, respectively) and from A. baumannii ATCC 17978 transformed with OMVs from clinic isolates AbH12O-A2 and AbH12O-CU3 (blots C and D, respectively). The mock-transformed A. baumannii strain ATCC 17978 (blot E) was used as a negative control. Equal amounts of OMVs of each sample were analyzed in the dot blots.
Fig. 4.
REP-PCR profile of the A. baumannii strains used in this study. Blot A, AbH12O-A2; blot B, AbH12O-CU3; blot C, ATCC 17978; blot D, ATCC 17978 transformed with OMVs from AbH12O-A2; blot E, ATCC 17978 transformed with OMVs from AbH12O-CU3.
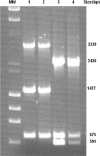
Fig. 5.
PCR fingerprinting profile of plasmids pMMA2 and pMCU3, determined by using a set of oligonucleotides. Lane 1, PCR fingerprint of plasmids isolated from strain AbH12O-A2; lane 2, PCR fingerprint of plasmids isolated from strain ATCC 17978 transformed with OMVs from AbH12O-A2; lane 3, PCR fingerprint of plasmids isolated from strain AbH12O-CU3; lane 4, PCR fingerprint of plasmids isolated from strain ATCC 17978 transformed with OMVs from AbH12O-CU3 as a source. MW, λ DNA-HindIII and φ174 DNA-HaeIII mix.
Similar articles
- Carbapenem Resistance in Acinetobacter nosocomialis and Acinetobacter junii Conferred by Acquisition of _bla_OXA-24/40 and Genetic Characterization of the Transmission Mechanism between Acinetobacter Genomic Species.
Lasarte-Monterrubio C, Guijarro-Sánchez P, Bellés A, Vázquez-Ucha JC, Arca-Suárez J, Fernández-Lozano C, Bou G, Beceiro A; Spanish National Study Acinetobacter spp. 2020 Group. Lasarte-Monterrubio C, et al. Microbiol Spectr. 2022 Feb 23;10(1):e0273421. doi: 10.1128/spectrum.02734-21. Epub 2022 Feb 9. Microbiol Spectr. 2022. PMID: 35138195 Free PMC article. - [Investigation of OXA type beta-lactamases and PFGE patterns in Acinetobacter baumannii strains resistant to carbapenems].
Keyik S, Arslan U, Türk Dağı H, Seyhan T, Fındık D. Keyik S, et al. Mikrobiyol Bul. 2014 Oct;48(4):556-65. doi: 10.5578/mb.8274. Mikrobiyol Bul. 2014. PMID: 25492651 Turkish. - Prevalence of different carbapenemase genes among carbapenem-resistant Acinetobacter baumannii blood isolates in Taiwan.
Wang TH, Leu YS, Wang NY, Liu CP, Yan TR. Wang TH, et al. Antimicrob Resist Infect Control. 2018 Oct 11;7:123. doi: 10.1186/s13756-018-0410-5. eCollection 2018. Antimicrob Resist Infect Control. 2018. PMID: 30338061 Free PMC article. - Carbapenem resistance in Acinetobacter baumannii: mechanisms and epidemiology.
Poirel L, Nordmann P. Poirel L, et al. Clin Microbiol Infect. 2006 Sep;12(9):826-36. doi: 10.1111/j.1469-0691.2006.01456.x. Clin Microbiol Infect. 2006. PMID: 16882287 Review. - OXA β-lactamases.
Evans BA, Amyes SG. Evans BA, et al. Clin Microbiol Rev. 2014 Apr;27(2):241-63. doi: 10.1128/CMR.00117-13. Clin Microbiol Rev. 2014. PMID: 24696435 Free PMC article. Review.
Cited by
- Pathogenesis Mediated by Bacterial Membrane Vesicles.
Gilmore WJ, Bitto NJ, Kaparakis-Liaskos M. Gilmore WJ, et al. Subcell Biochem. 2021;97:101-150. doi: 10.1007/978-3-030-67171-6_6. Subcell Biochem. 2021. PMID: 33779916 - Mycoplasmas and Their Antibiotic Resistance: The Problems and Prospects in Controlling Infections.
Chernova OA, Medvedeva ES, Mouzykantov AA, Baranova NB, Chernov VM. Chernova OA, et al. Acta Naturae. 2016 Apr-Jun;8(2):24-34. Acta Naturae. 2016. PMID: 27437137 Free PMC article. - Virulence Characteristics and Emerging Therapies for Biofilm-Forming Acinetobacter baumannii: A Review.
Dolma KG, Khati R, Paul AK, Rahmatullah M, de Lourdes Pereira M, Wilairatana P, Khandelwal B, Gupta C, Gautam D, Gupta M, Goyal RK, Wiart C, Nissapatorn V. Dolma KG, et al. Biology (Basel). 2022 Sep 12;11(9):1343. doi: 10.3390/biology11091343. Biology (Basel). 2022. PMID: 36138822 Free PMC article. Review. - Membrane Vesicles of Clostridioides difficile and Other Clostridial Species.
Goh S, Inal J. Goh S, et al. Adv Exp Med Biol. 2024;1435:315-327. doi: 10.1007/978-3-031-42108-2_14. Adv Exp Med Biol. 2024. PMID: 38175481 - Comparative Genomics Analysis and Outer Membrane Vesicle-Mediated Horizontal Antibiotic-Resistance Gene Transfer in Avibacterium paragallinarum.
Xu J, Mei C, Zhi Y, Liang ZX, Zhang X, Wang HJ. Xu J, et al. Microbiol Spectr. 2022 Oct 26;10(5):e0137922. doi: 10.1128/spectrum.01379-22. Epub 2022 Aug 24. Microbiol Spectr. 2022. PMID: 36000914 Free PMC article.
References
- Bou G., Cervero G., Dominguez M. A., Quereda C., Martinez-Beltran J. 2000. PCR-based DNA fingerprinting (REP-PCR, AP-PCR) and pulsed-field gel electrophoresis characterization of a nosocomial outbreak caused by imipenem- and meropenem-resistant Acinetobacter baumannii. Clin. Microbiol. Infect. 6:635–643 - PubMed
- Bradford M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248–254 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources